NICGAR: a Niching Genetic Algorithm to Mine a Diverse Set of Interesting Quantitative Association Rules
This Website contains complementary material to the paper:
D. Martín, J. Alcalá-Fdez, A. Rosete and F.Herrera, NICGAR: a Niching Genetic Algorithm to Mine a Diverse Set of Interesting Quantitative Association Rules. Information Sciences 355-356 (2016) 208-228. DOI: 10.1016/j.ins.2016.03.039
Summary:
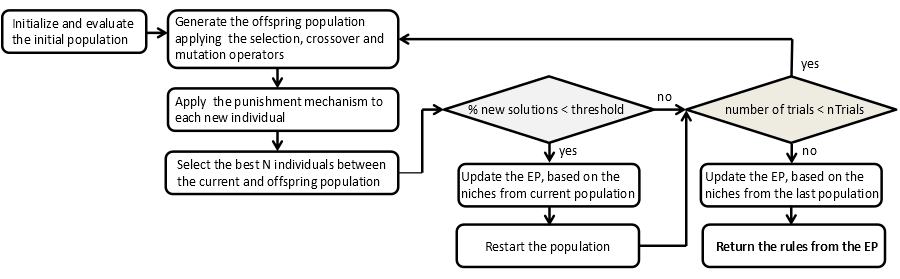
Flowchart of the algorithm
NICGAR can be summarized as follows:
- Input: Population size N, number of evaluations nTrials, probability of mutation Pmut, factor of amplitude for each attribute of the dataset ρ, difference threshold α, niching threshold NichMin, quality threshold EvMin.
- Output: External population (EP)
- Initialize
- Generate the initial population (Pt) with N chromosomes.
- Evaluate the initial population.
- Generate the offspring population (Qt) as follows.
- Two parent individuals x1 and x2 are selected by binary tournament selection using the fitness function.
- Generate two offspring applying the crossover operator on the individuals x1 and x2. Next, the offspring are mutated and repaired using the appropriate operators.
- The offspring is evaluated.
- If the EP is not empty, the punishment mechanism is applied to each new individual if they belong to a niche from the EP and their fitness is lower than its corresponding solution of the EP.
- Join the parents with their offspring and select the best N individuals to take part in the next population (Pt+1).
- If the difference between the current population and the previous population is less than α%:
- Update the EP, based on the niches from Pt+1.
- Restart the population and apply the punishment mechanism to the new individuals that have been created in the restarting process.
- If the maximum number of evaluations is not reached, go to Step 2.
- Update the EP, based on the niches from the last population.
- The EP is returned.
A scheme of this algorithm is shown in the Figure 1.
Figure 1. Scheme of the algorithm NICGAR.
Experimental Framework
Data-sets used in the paper
Table 1 describes the 27 real-world datasets that have been used in this experimental study, where "Attributes(R/I/N)" is the number of attributes (Real/Integer/Nominal) in the data, "Examples" is the number of examples and "Download" contains a link for downloading each data-set in the KEEL format. You may also download all data-sets by clicking here. Moreover, the datasets are available in the repository KEEL-dataset, where they can be downloaded (available at KEEL-dataset Repository).
Table 1: Summary of the datasets considered for the experimental study
Algorithms analyzed
In these experiments, we compare the proposed approach with seven other algorithms, which are available from the KEEL software tool. A brief description of these algorithms follows.
- Clearing: This Niching Genetic Algorithm is one of the most popular niching techniques used in conjunction with the evolutionary computation community. This method is based on limited the resources within subpopulations of similar individuals, where only the best individuals of each niche will survive since the fitness of the rest of them will be reset to zero. This process is applied after evaluation process and before the selection.
- Adaptive Species Conservation Genetic Algorithm (ASCGA): This algorithm is based on the species conservation approach (SCGA) proposed by Li, it divides the population into several species according to their similarity and each of these species is built around a dominating individual, commonly referred to as a species seed. This method automatically adjust species parameters and allow the species to adapt to an optimization problem.
- Evolutionary Association Rules Mining with Genetic Algorithm (EARMGA): This algorithm uses a GA to identify QARs. Each chromosome encodes a generalized k-rule, where k indicates the desired length. The most interesting rules are returned according to the interestingness measure defined by the fitness function, which is based on the support of the rule and its antecedent and consequent support. The proposed GA performs a dataset-independent approach that does not rely upon the minSup threshold.
- GENetic Association Rules(GENAR): This algorithm uses a GA to mine association rules in numeric datasets. Each chromosome encodes an association rule, containing maximum and minimum intervals of each numeric attribute. The length of the rules is always fixed to the number of attributes, only the last attribute forms the consequent. The objective function considers the number of records covered by the rule and penalizes those which have already covered the same records in the dataset.
- Genetic Association Rules (GAR): This algorithm is an extension of GENAR, which searches for frequent itemsets in numeric datasets without needing to discretize the attributes. Each chromosome is a k-itemset, in which each gene represents the maximum and minimum values of the attributes that belong to the k-itemset. This algorithm finds frequent itemsets, and it is therefore necessary to run another procedure afterwards in order to generate association rules.
- Genetic algorithm for automated mining of both positive and negative quantitative association rules (Alatasetal): This algorithm designs a GA, which does not require a user-specified the minSup and minConf thresholds, to simultaneously search for intervals of quantitative attributes and to discover PNQARs that these intervals conform to in a single run. The chromosomes represent rules, in which each gene has four parts.
- QAR-CIP-NSGA-II: This method mines a set of QARs with a good trade-off between interpretability and accuracy, which maximizes three objectives: comprehensibility, interestingness and performance, understanding by performance the product of Certainty Factor (CF) and support. This algorithm extends the well-known MOEA NSGA-II to perform an evolutionary learning of the intervals of the attributes and a condition selection for each rule.
- MOPNAR: This algorithm extends the MOEA based on decomposition MOEA/D-DE to obtain a reduced set of PNQARs with a good trade-off between the number of rules, support and coverage of the dataset. It performs a condition selection and an evolutionary learning of the intervals of the attributes for each rule, maximizing three objectives: comprehensibility; interestingness and performance.
- Apriori: Apriori follows a breadth-first strategy. It generates candidate itemsets for the current iteration by means of itemsets considered to be frequent in the previous iteration. It then enumerates all the subsets for each transaction and increments the support of candidates matching them. Then, those that have the user-specified minSup are marked as frequent for the next iteration. This process is repeated until all frequent itemsets have been found. Finally, Apriori uses the frequent itemsets to generate positive rules with confidence greater than minimum confidence.
- Eclat: Eclat employs a depth-first strategy. It generates candidates by extending the prefixes of an itemset until an infrequent one is found. In such cases, it simply backtracks to the previous prefix and then recursively applies the above procedure. Unlike Apriori, for all the items in a dataset, it first constructs a list of all the transaction identifiers (tid-list) containing that item. Then it counts the support by merely intersecting two or more tid-lists to check whether they have items in common. If so, the support is equal to the size of the resulting set. The process for generating the positive rules is the same as Apriori.
Results obtained
This section is divided into three parts: the first part provides the results of the comparison with two NGAs extended to extract PNQARs. Next, the second part shows the results of the comparison with other mono-objective and multi-objective evolutionary approaches. Finally, in the third part, the performance results of our algorithm again two classical algorithms can be found.
Results obtained in the comparison with other NGAs
In this section, we present the performance results obtained of our algorithm against Clearing and ASCGA. Table 2 shows the average results obtained of 5 runs for each dataset.
Moreover, this performance results are available for download in xls, pdf and tex files by clicking the links ![]() ,
, ![]() and
and ![]() , respectively.
, respectively.
| Algorithm | #R | AvSup(s) | AvConf(s) | AvLift (s) | AvConv | AvCF (s) | AvNetconf(s) | AvYulesQ (s) | AvAmp (s) | AvDiv (s) | %Trans (s) |
| Balance | |||||||||||
| Clearing | 24.20 | 0.08 (0.05) | 0.82 (0.10) | 2.74 (1.15) | 8 | 0.68 (0.21) | 0.49 (0.18) | 0.73 (0.17) | 3.47 (0.94) | 0.73 (0.04) | 89.48 (10.20) |
| ASCGA | 21.20 | 0.09 (0.03) | 0.73 (0.05) | 2.30 (0.44) | 8 | 0.54 (0.07) | 0.39 (0.08) | 0.62 (0.09) | 3.34 (0.24) | 0.74 (0.05) | 75.01 (21.02) |
| NICGAR | 26.40 | 0.06 (0.02) | 0.92 (0.03) | 4.25 (0.44) | 8 | 0.86 (0.04) | 0.69 (0.03) | 0.91 (0.02) | 3.81 (0.18) | 0.82 (0.02) | 89.96 (2.19) |
| Basketball | |||||||||||
| Clearing | 57.00 | 0.03 (0.01) | 0.98 (0.02) | 32.87 (9.48) | 8 | 0.97 (0.03) | 0.91 (0.03) | 0.99 (0.01) | 2 (0.01) | 0.99 (0.00) | 84.17 (8.12) |
| ASCGA | 10.00 | 0.09 (0.07) | 0.97 (0.03) | 14.81 (11.66) | 8 | 0.94 (0.05) | 0.83 (0.07) | 0.98 (0.02) | 2.04 (0.05) | 0.81 (0.16) | 35.84 (9.84) |
| NICGAR | 49.80 | 0.04 (0.01) | 0.99 (0.01) | 9.43 (2.53) | 8 | 0.98 (0.02) | 0.80 (0.04) | 0.99 (0.01) | 2.15 (0.09) | 0.95 (0.01) | 91.67 (4.04) |
| Bolts | |||||||||||
| Clearing | 21.80 | 0.13 (0.07) | 0.96 (0.04) | 9.50 (4.16) | 8 | 0.94 (0.05) | 0.84 (0.05) | 0.98 (0.02) | 2.06 (0.10) | 0.87 (0.06) | 99.00 (1.37) |
| ASCGA | 15.60 | 0.18 (0.04) | 0.96 (0.03) | 4.91 (2.43) | 8 | 0.93 (0.05) | 0.82 (0.06) | 0.97 (0.03) | 2.42 (0.29) | 0.74 (0.10) | 89.00 (16.73) |
| NICGAR | 9.80 | 0.30 (0.04) | 0.98 (0.02) | 5.51 (1.66) | 8 | 0.98 (0.02) | 0.93 (0.03) | 1 (0.00) | 2 (0.00) | 0.82 (0.02) | 100.00 (0.00) |
| Coil2000 | |||||||||||
| Clearing | 18.60 | 0.23 (0.10) | 0.80 (0.06) | 5.93 (5.20) | 8 | 0.69 (0.08) | 0.63 (0.05) | 0.85 (0.06) | 2.08 (0.12) | 0.75 (0.04) | 99.81 (0.36) |
| ASCGA | 29.00 | 0.22 (0.07) | 0.63 (0.02) | 4.21 (5.46) | 8 | 0.38 (0.05) | 0.32 (0.05) | 0.54 (0.04) | 2.71 (0.26) | 0.81 (0.03) | 99.62 (0.82) |
| NICGAR | 25.40 | 0.28 (0.03) | 0.93 (0.02) | 3.65 (0.51) | 8 | 0.89 (0.03) | 0.86 (0.03) | 0.98 (0.01) | 2.04 (0.04) | 0.82 (0.03) | 99.96 (0.10) |
| Fars | |||||||||||
| Clearing | 21.60 | 0.30 (0.07) | 0.93 (0.02) | 12.92 (16.92) | 8 | 0.87 (0.04) | 0.85 (0.04) | 0.93 (0.05) | 2.20 (0.04) | 0.68 (0.04) | 92.87 (15.06) |
| ASCGA | 22.60 | 0.22 (0.05) | 0.78 (0.09) | 8.11 (8.17) | 8 | 0.61 (0.12) | 0.56 (0.11) | 0.71 (0.14) | 2.51 (0.21) | 0.79 (0.02) | 99.48 (1.17) |
| NICGAR | 12.60 | 0.31 (0.05) | 0.99 (0.00) | 5.67 (0.58) | 8 | 0.99 (0.01) | 0.98 (0.02) | 1 (0.00) | 2.02 (0.04) | 0.81 (0.04) | 100.00 (0.00) |
| House16H | |||||||||||
| Clearing | 24.60 | 0.09 (0.06) | 0.81 (0.05) | 13.12 (8.20) | 8 | 0.74 (0.04) | 0.69 (0.06) | 0.55 (0.22) | 2.09 (0.07) | 0.72 (0.09) | 83.41 (21.75) |
| ASCGA | 11.60 | 0.14 (0.06) | 0.75 (0.13) | 14.42 (24.64) | 8 | 0.63 (0.19) | 0.57 (0.20) | 0.70 (0.07) | 2.27 (0.31) | 0.70 (0.08) | 68.69 (16.64) |
| NICGAR | 6.00 | 0.24 (0.04) | 0.92 (0.00) | 3.67 (0.71) | 15.00 | 0.88 (0.01) | 0.83 (0.01) | 0.98 (0.00) | 2 (0.00) | 0.86 (0.02) | 82.18 (13.91) |
| Ionosphere | |||||||||||
| Clearing | 41.00 | 0.07 (0.02) | 0.94 (0.02) | 55.85 (25.71) | 8 | 0.91 (0.03) | 0.86 (0.05) | 0.98 (0.01) | 2.01 (0.02) | 0.92 (0.03) | 92.03 (4.34) |
| ASCGA | 13.00 | 0.18 (0.04) | 0.78 (0.07) | 3.13 (0.64) | 8 | 0.67 (0.11) | 0.59 (0.09) | 0.88 (0.08) | 2.43 (0.13) | 0.82 (0.07) | 82.51 (17.39) |
| NICGAR | 24.80 | 0.20 (0.02) | 0.85 (0.01) | 3.77 (0.59) | 8 | 0.78 (0.02) | 0.74 (0.02) | 0.96 (0.00) | 2.01 (0.02) | 0.88 (0.01) | 99.83 (0.15) |
| Letter | |||||||||||
| Clearing | 17.40 | 0.16 (0.03) | 0.87 (0.03) | 7.28 (1.87) | 8 | 0.77 (0.03) | 0.72 (0.05) | 0.93 (0.01) | 2.11 (0.08) | 0.81 (0.04) | 98.26 (1.59) |
| ASCGA | 15.00 | 0.18 (0.09) | 0.74 (0.06) | 4.61 (0.76) | 8 | 0.59 (0.08) | 0.57 (0.09) | 0.82 (0.07) | 2.40 (0.25) | 0.76 (0.07) | 85.47 (24.98) |
| NICGAR | 18.80 | 0.15 (0.02) | 0.89 (0.02) | 5.94 (1.31) | 8 | 0.84 (0.03) | 0.76 (0.04) | 0.96 (0.01) | 2.01 (0.02) | 0.87 (0.01) | 98.17 (1.62) |
| Magic | |||||||||||
| Clearing | 9.80 | 0.34 (0.18) | 0.93 (0.01) | 2.81 (0.64) | 10.38 | 0.79 (0.24) | 0.85 (0.03) | 0.99 (0.01) | 2.01 (0.02) | 0.77 (0.16) | 99.55 (0.32) |
| ASCGA | 9.20 | 0.22 (0.09) | 0.89 (0.06) | 2.88 (0.35) | 14.25 | 0.82 (0.08) | 0.72 (0.05) | 0.92 (0.07) | 2.02 (0.05) | 0.57 (0.18) | 69.57 (15.76) |
| NICGAR | 6.60 | 0.26 (0.01) | 0.94 (0.01) | 3.00 (0.06) | 15.30 | 0.91 (0.02) | 0.85 (0.01) | 0.99 (0.01) | 2 (0.00) | 0.85 (0.01) | 94.87 (4.93) |
| Movement Libras | |||||||||||
| Clearing | 23.60 | 0.24 (0.01) | 0.94 (0.03) | 5.11 (2.50) | 8 | 0.90 (0.04) | 0.88 (0.04) | 0.98 (0.02) | 2 (0.00) | 0.87 (0.01) | 99.95 (0.12) |
| ASCGA | 20.80 | 0.21 (0.06) | 0.82 (0.11) | 2.81 (0.65) | 8 | 0.70 (0.18) | 0.63 (0.21) | 0.83 (0.15) | 2.57 (0.51) | 0.84 (0.05) | 94.73 (11.34) |
| NICGAR | 27.80 | 0.27 (0.01) | 0.98 (0.00) | 3.64 (0.05) | 8 | 0.97 (0.01) | 0.95 (0.01) | 1.00 (0.00) | 2 (0.00) | 0.86 (0.00) | 100.00 (0.00) |
| Optdigits | |||||||||||
| Clearing | 17.80 | 0.22 (0.12) | 0.84 (0.06) | 5.05 (3.47) | 8 | 0.69 (0.14) | 0.65 (0.07) | 0.84 (0.05) | 2.07 (0.16) | 0.76 (0.07) | 96.82 (4.63) |
| ASCGA | 23.80 | 0.23 (0.07) | 0.67 (0.04) | 2.07 (0.55) | 8 | 0.40 (0.08) | 0.37 (0.04) | 0.60 (0.07) | 2.44 (0.14) | 0.80 (0.03) | 98.99 (1.06) |
| NICGAR | 28.80 | 0.28 (0.02) | 0.87 (0.02) | 3.24 (0.40) | 8 | 0.75 (0.05) | 0.71 (0.01) | 0.95 (0.01) | 2.05 (0.04) | 0.82 (0.02) | 100.00 (0.00) |
| Penbased | |||||||||||
| Clearing | 16.80 | 0.15 (0.04) | 0.82 (0.05) | 4.22 (2.14) | 8 | 0.73 (0.08) | 0.63 (0.08) | 0.87 (0.05) | 2.06 (0.06) | 0.84 (0.02) | 98.08 (1.10) |
| ASCGA | 19.80 | 0.22 (0.04) | 0.76 (0.02) | 2.20 (0.31) | 8 | 0.60 (0.05) | 0.53 (0.04) | 0.82 (0.05) | 2.12 (0.06) | 0.66 (0.20) | 94.04 (6.20) |
| NICGAR | 15.00 | 0.18 (0.02) | 0.89 (0.01) | 3.64 (0.77) | 8 | 0.83 (0.02) | 0.73 (0.03) | 0.95 (0.01) | 2.06 (0.10) | 0.88 (0.02) | 98.64 (0.92) |
| Pollution | |||||||||||
| Clearing | 48.60 | 0.05 (0.01) | 0.98 (0.03) | 18.68 (3.55) | 8 | 0.97 (0.04) | 0.90 (0.04) | 0.99 (0.03) | 2.01 (0.02) | 0.96 (0.01) | 90.34 (8.53) |
| ASCGA | 13.00 | 0.13 (0.07) | 0.95 (0.05) | 9.54 (3.87) | 8 | 0.92 (0.06) | 0.83 (0.06) | 0.98 (0.03) | 2.16 (0.23) | 0.85 (0.10) | 72.67 (22.69) |
| NICGAR | 57.80 | 0.08 (0.01) | 0.98 (0.01) | 8.35 (1.33) | 8 | 0.97 (0.01) | 0.84 (0.02) | 1.00 (0.00) | 2.07 (0.05) | 0.93 (0.01) | 100.00 (0.00) |
| Quake | |||||||||||
| Clearing | 33.20 | 0.03 (0.03) | 0.86 (0.08) | 38.81 (28.61) | 8 | 0.84 (0.09) | 0.76 (0.11) | 0.84 (0.13) | 2 (0.00) | 0.88 (0.06) | 75.42 (14.14) |
| ASCGA | 4.60 | 0.28 (0.13) | 0.84 (0.06) | 3.39 (2.64) | 5.64 | 0.72 (0.12) | 0.66 (0.07) | 0.87 (0.08) | 2.08 (0.18) | 0.62 (0.12) | 72.51 (25.18) |
| NICGAR | 3.60 | 0.28 (0.05) | 0.90 (0.01) | 2.51 (0.80) | 11.40 | 0.81 (0.02) | 0.72 (0.02) | 0.95 (0.01) | 2 (0.00) | 0.75 (0.03) | 83.15 (1.70) |
| Satimage | |||||||||||
| Clearing | 15.20 | 0.25 (0.03) | 0.92 (0.02) | 5.00 (3.32) | 8 | 0.86 (0.03) | 0.80 (0.03) | 0.98 (0.01) | 2.01 (0.03) | 0.82 (0.01) | 96.34 (6.91) |
| ASCGA | 30.60 | 0.37 (0.04) | 0.83 (0.05) | 2.69 (0.10) | 8 | 0.66 (0.05) | 0.66 (0.06) | 0.93 (0.01) | 2.39 (0.15) | 0.71 (0.02) | 100.00 (0.00) |
| NICGAR | 24.00 | 0.24 (0.01) | 0.92 (0.01) | 3.83 (0.31) | 12.27 | 0.88 (0.01) | 0.85 (0.01) | 0.99 (0.00) | 2 (0.00) | 0.82 (0.01) | 93.94 (2.29) |
| Segment | |||||||||||
| Clearing | 11.80 | 0.20 (0.01) | 0.93 (0.05) | 4.41 (0.80) | 8 | 0.91 (0.07) | 0.88 (0.08) | 0.94 (0.09) | 2.02 (0.04) | 0.86 (0.03) | 98.09 (3.04) |
| ASCGA | 16.00 | 0.25 (0.06) | 0.86 (0.05) | 3.38 (0.69) | 8 | 0.79 (0.05) | 0.74 (0.10) | 0.88 (0.08) | 2.25 (0.18) | 0.77 (0.05) | 98.76 (1.42) |
| NICGAR | 12.60 | 0.22 (0.01) | 0.99 (0.01) | 4.91 (0.43) | 8 | 0.98 (0.01) | 0.97 (0.01) | 1.00 (0.00) | 2 (0.00) | 0.88 (0.01) | 99.76 (0.26) |
| Sonar | |||||||||||
| Clearing | 51.40 | 0.06 (0.03) | 0.96 (0.02) | 57.68 (14.37) | 8 | 0.93 (0.03) | 0.91 (0.03) | 0.99 (0.01) | 2 (0.00) | 0.94 (0.03) | 87.02 (22.84) \\ |
| ASCGA | 18.20 | 0.20 (0.06) | 0.77 (0.11) | 4.12 (2.92) | 8 | 0.58 (0.19) | 0.51 (0.18) | 0.74 (0.17) | 2.57 (0.43) | 0.85 (0.04) | 95.48 (6.32) \\ |
| NICGAR | 22.80 | 0.26 (0.03) | 0.89 (0.02) | 7.56 (5.59) | 8 | 0.81 (0.02) | 0.77 (0.02) | 0.97 (0.01) | 2.01 (0.02) | 0.84 (0.02) | 100.00 (0.00) |
| Spambase | |||||||||||
| Clearing | 32.40 | 0.09 (0.12) | 0.89 (0.05) | 115.83 (93.33) | 8 | 0.83 (0.09) | 0.81 (0.10) | 0.88 (0.09) | 2.03 (0.04) | 0.84 (0.14) | 85.01 (23.67) |
| ASCGA | 20.20 | 0.19 (0.03) | 0.75 (0.06) | 20.22 (20.15) | 8 | 0.59 (0.04) | 0.45 (0.10) | 0.72 (0.08) | 2.72 (0.33) | 0.78 (0.04) | 95.85 (6.84) |
| NICGAR | 11.20 | 0.12 (0.02) | 0.95 (0.02) | 73.02 (28.16) | 8 | 0.90 (0.03) | 0.87 (0.04) | 0.98 (0.01) | 2 (0.00) | 0.84 (0.05) | 76.75 (31.96) |
| Spectfheart | |||||||||||
| Clearing | 39.00 | 0.12 (0.08) | 0.97 (0.02) | 61.20 (11.76) | 8 | 0.91 (0.06) | 0.94 (0.03) | 0.99 (0.01) | 2.01 (0.01) | 0.86 (0.06) | 86.22 (30.82) |
| ASCGA | 12.60 | 0.37 (0.10) | 0.89 (0.03) | 18.04 (10.86) | 8 | 0.68 (0.10) | 0.80 (0.07) | 0.95 (0.03) | 2.17 (0.19) | 0.71 (0.11) | 99.25 (1.09) |
| NICGAR | 72.20 | 0.13 (0.01) | 0.98 (0.00) | 29.01 (4.73) | 8 | 0.92 (0.00) | 0.92 (0.02) | 1.00 (0.00) | 2.03 (0.05) | 0.89 (0.02) | 100.00 (0.00) |
| Stock | |||||||||||
| Clearing | 14.60 | 0.21 (0.07) | 0.91 (0.05) | 16.56 (18.61) | 8 | 0.86 (0.07) | 0.78 (0.04) | 0.96 (0.04) | 2.06 (0.10) | 0.84 (0.05) | 99.41 (0.98) |
| ASCGA | 8.00 | 0.28}(0.09) | 0.90 (0.06) | 2.70 (0.46) | 8 | 0.85 (0.09) | 0.79 (0.11) | 0.95 (0.05) | 2.18 (0.24) | 0.70 (0.13) | 81.04 (11.23) |
| NICGAR | 9.20 | 0.28}(0.02) | 0.95 (0.01) | 3.01 (0.18) | 8 | 0.93 (0.02) | 0.87 (0.01) | 0.99 (0.00) | 2.07 (0.07) | 0.86 (0.02) | 99.41 (0.23) |
| Stulong | |||||||||||
| Clearing | 10.60 | 0.23 (0.12) | 0.87 (0.02) | 29.51 (43.04) | 8 | 0.68 (0.10) | 0.75 (0.11) | 0.94 (0.04) | 2.10 (0.14) | 0.72 (0.10) | 97.20 (1.52) |
| ASCGA | 7.40 | 0.16 (0.03) | 0.79 (0.01) | 9.08 (7.99) | 3.44 | 0.71 (0.02) | 0.65 (0.01) | 0.92 (0.01) | 2 (0.00) | 0.45 (0.05) | 44.93 (23.77) |
| NICGAR | 4.20 | 0.26 (0.07) | 0.79 (0.03) | 16.82 (19.90) | 8 | 0.62 (0.08) | 0.64 (0.06) | 0.90 (0.03) | 2.04 (0.09) | 0.78 (0.06) | 84.99 (7.01) |
| Texture | |||||||||||
| Clearing | 15.20 | 0.29 (0.04) | 0.93 (0.02) | 3.14 (0.32) | 19.47 | 0.89 (0.03) | 0.86 (0.04) | 0.98 (0.02) | 2.01 (0.03) | 0.83 (0.02) | 97.26 (5.77) |
| ASCGA | 16.40 | 0.29 (0.04) | 0.90 (0.04) | 35.96 (56.31) | 8 | 0.81 (0.07) | 0.75 (0.06) | 0.93 (0.06) | 2.37 (0.28) | 0.76 (0.09) | 98.23 (2.05) |
| NICGAR | 17.40 | 0.27 (0.02) | 0.97 (0.00) | 3.72 (0.39) | 8 | 0.95 (0.00) | 0.92 (0.01) | 1.00 (0.00) | 2 (0.00) | 0.82 (0.02) | 95.85 (4.66) |
| Thyroid | |||||||||||
| Clearing | 27.00 | 0.11 (0.16) | 0.87 (0.08) | 16.05 (9.81) | 8 | 0.76 (0.21) | 0.77 (0.08) | 0.81 (0.10) | 2.48 (0.38) | 0.73 (0.12) | 72.81 (40.62) |
| ASCGA | 20.20 | 0.17 (0.07) | 0.74 (0.06) | 6.14 (4.15) | 8 | 0.53 (0.11) | 0.38 (0.16) | 0.56 (0.16) | 2.64 (0.43) | 0.71 (0.09) | 88.64 (13.24) |
| NICGAR | 6.20 | 0.23 (0.07) | 0.93 (0.02) | 14.29 (9.46) | 8 | 0.80 (0.07) | 0.85 (0.02) | 0.95 (0.05) | 2.02 (0.05) | 0.79 (0.05) | 98.16 (2.30) |
| Wdbc | |||||||||||
| Clearing | 18.80 | 0.19 (0.05) | 0.92 (0.04) | 48.87 (36.99) | 8 | 0.88 (0.06) | 0.84 (0.07) | 0.97 (0.03) | 2 (0.00) | 0.86 (0.01) | 99.51 (0.62) |
| ASCGA | 13.80 | 0.26 (0.04) | 0.81 (0.08) | 3.18 (1.52) | 8 | 0.68 (0.14) | 0.62 (0.17) | 0.81 (0.13) | 2.37 (0.35) | 0.78 (0.10) | 97.62 (2.44) |
| NICGAR | 13.60 | 0.27 (0.03) | 0.96 (0.02) | 3.49 (0.39) | 8 | 0.93 (0.04) | 0.90 (0.03) | 0.99 (0.01) | 2 (0.00) | 0.84 (0.01) | 98.53 (2.10) |
| Vehicle | |||||||||||
| Clearing | 13.00 | 0.26 (0.05) | 0.96 (0.02) | 5.21 (3.69) | 8 | 0.93 (0.04) | 0.89 (0.04) | 0.97 (0.04) | 2 (0.00) | 0.84 (0.03) | 99.95 (0.10) |
| ASCGA | 13.80 | 0.25 (0.03) | 0.92 (0.05) | 3.35 (0.57) | 8 | 0.87 (0.07) | 0.84 (0.08) | 0.96 (0.04) | 2.08 (0.08) | 0.76 (0.05) | 96.86 (2.81) |
| NICGAR | 12.80 | 0.27 (0.03) | 0.97 (0.01) | 3.46 (0.26) | 8 | 0.95 (0.02) | 0.92 (0.01) | 1.00 (0.00) | 2 (0.00) | 0.86 (0.02) | 99.98 (0.05) |
| Wine | |||||||||||
| Clearing | 23.20 | 0.14 (0.10) | 0.94 (0.07) | 19.43 (18.27) | 8 | 0.91 (0.09) | 0.83 (0.11) | 0.97 (0.05) | 2.02 (0.03) | 0.92 (0.09) | 93.49 (7.39) |
| ASCGA | 8.40 | 0.25 (0.04) | 0.88 (0.06) | 3.70 (2.98) | 8 | 0.79 (0.11) | 0.73 (0.11) | 0.92 (0.07) | 2.17 (0.25) | 0.78 (0.07) | 83.71 (21.15) |
| NICGAR | 6.60 | 0.28 (0.01) | 0.94 (0.01) | 2.79 (0.18) | 8 | 0.90 (0.02) | 0.84 (0.02) | 0.99 (0.01) | 2 (0.00) | 0.86 (0.00) | 94.61 (0.85) |
| Vowel | |||||||||||
| Clearing | 51.60 | 0.23 (0.40) | 0.96 (0.02) | 92.68 (69.98) | 8 | 0.78 (0.33) | 0.91 (0.04) | 0.50 (0.15) | 2.02 (0.02) | 0.81 (0.21) | 92.10 (8.89) |
| ASCGA | 15.40 | 0.19 (0.07) | 0.79 (0.07) | 4.41 (3.00) | 8 | 0.68 (0.12) | 0.59 (0.12) | 0.81 (0.11) | 2.40 (0.22) | 0.77 (0.08) | 92.89 (12.00) |
| NICGAR | 8.40 | 0.27 (0.11) | 0.95 (0.05) | 5.81 (2.57) | 8 | 0.92 (0.07) | 0.89 (0.08) | 0.95 (0.09) | 2.02 (0.05) | 0.85 (0.01) | 100.00 (0.00) |
Results obtained in the comparison with mono-objective and multi-objective evolutionary approaches
In this section, we present the results obtained by our algorithm against four mono-objective evolutionary algorithms (EARMGA, GAR, GENAR and Alatasetal) and two multi-objective evolutionary algorithms (QAR-CIP-NSGA-II and MOPNAR). Table 3 and Table 4 show the average results obtained by the mono-objectives and the multi-objectives evolutionary approaches of 5 runs for each dataset, respectively.
Moreover, this performance results are available for download in xls, pdf and tex files. To download the results obtained by the mono-objective evolutionary algoritmhs, click the links by clicking the links ![]() ,
, ![]() and
and ![]() and for the ones obtained by the multi-objective evolutionary algorithms, click the links
and for the ones obtained by the multi-objective evolutionary algorithms, click the links ![]() ,
, ![]() and
and ![]() .
.
| Algorithm | #R | AvSup (σ) | AvConf (σ) | AvLift (σ) | AvConv | AvCF (σ) | AvNetconf (σ) | AvYulesQ (σ) | AvAmp (σ) | AvDiv (σ) | %Trans (σ) |
| Balance | |||||||||||
| EARMGA | 100 | 0.5 (0.04) | 1 (0) | 1 (0) | 1 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.46 (0.02) | 100 (0) |
| GENAR | 30 | 0.13 (0) | 0.94 (0.01) | 2.04 (0.02) | 31.04 | 0.89 (0.01) | 0.55 (0.01) | 0.92 (0.01) | 5 (0) | 0.6 (0.01) | 85.64 (0.96) |
| GAR | 0 | - | - | - | - | - | - | - | - | - | - |
| Alatasetal | 24.4 | 0.31 (0.07) | 1 (0) | 1.1 (0.12) | ∞ | 0.06 (0.05) | 0.03 (0.02) | 0.05 (0.05) | 4.35 (0.49) | 0.48 (0.04) | 100 (0) |
| NICGAR | 26.4 | 0.06 (0.02) | 0.92 (0.03) | 4.25 (0.44) | ∞ | 0.86 (0.04) | 0.69 (0.03) | 0.91 (0.02) | 3.81 (0.18) | 0.82 (0.02) | 89.96 (2.19) |
| Basketball | |||||||||||
| EARMGA | 100 | 0.27 (0.04) | 1 (0) | 1.02 (0.02) | ∞ | 0.06 (0.08) | 0.01 (0.01) | 0.05 (0.05) | 2 (0) | 0.55 (0.02) | 100 (0) |
| GENAR | 30 | 0.3 (0.01) | 0.97 (0.01) | 1.12 (0.01) | ∞ | 0.7 (0.03) | 0.13 (0) | 0.67 (0.03) | 5 (0) | 0.3 (0.02) | 91.04 (2.81) |
| GAR | 2 | 0.75 (0.01) | 0.88 (0) | 1.02 (0) | 1.12 | 0.11 (0.01) | 0.11 (0.01) | 0.36 (0.04) | 2 (0) | 0 (0) | 96.88 (0) |
| Alatasetal | 8.6 | 0.98 (0.01) | 1 (0) | 1 (0) | 1 | -0.01 (0.01) | -0.01 (0.01) | 0 (0) | 3.2 (0.09) | 0.09 (0.09) | 100 (0) |
| NICGAR | 49.8 | 0.04 (0.01) | 0.99 (0.01) | 9.43 (2.53) | ∞ | 0.98 (0.02) | 0.8 (0.04) | 0.99 (0.01) | 2.15 (0.09) | 0.95 (0.01) | 91.67 (4.04) |
| Bolts | |||||||||||
| EARMGA | 100 | 0.34 (0.07) | 1 (0) | 1.05 (0.05) | ∞ | 0.15 (0.16) | 0.03 (0.04) | 0.15 (0.16) | 2 (0) | 0.57 (0.04) | 100 (0) |
| GENAR | 30 | 0.13 (0) | 1 (0) | 1.62 (0.07) | ∞ | 1 (0) | 0.42 (0.01) | 1 (0) | 8 (0) | 0.27 (0.05) | 44 (5.48) |
| GAR | 43 | 0.21 (0.03) | 0.99 (0.01) | 4.14 (0.3) | ∞ | 0.97 (0.01) | 0.88 (0.03) | 1 (0.01) | 3.31 (0.51) | 0.44 (0.07) | 81.5 (18.08) |
| Alatasetal | 21 | 0.95 (0.09) | 1 (0) | 1.04 (0.08) | ∞ | 0.14 (0.31) | 0.14 (0.31) | 0.14 (0.31) | 3.64 (0.43) | 0.02 (0.02) | 95 (8.66) |
| NICGAR | 9.8 | 0.3 (0.04) | 0.98 (0.02) | 5.51 (1.66) | ∞ | 0.98 (0.02) | 0.93 (0.03) | 1 (0) | 2 (0) | 0.82 (0.02) | 100 (0) |
| Coil2000 | |||||||||||
| EARMGA | 77 | 0.42 (0.17) | 1 (0) | 1.01 (0) | ∞ | 0.05 (0.04) | 0.01 (0) | 0 (0) | 2 (0) | 0.55 (0.09) | 100 (0) |
| GENAR | 30 | 0.01 (0) | 0.96 (0) | 1.02 (0.01) | ∞ | 0.34 (0.04) | 0.02 (0) | 0.01 (0.01) | 86 (0) | 0.53 (0) | 14.48 (0.6) |
| GAR | 197 | 0.94 (0.01) | 0.97 (0.01) | 1.01 (0) | ∞ | 0.04 (0.01) | 0.03 (0.01) | 0.08 (0.04) | 2.08 (0.03) | 0.37 (0.02) | 100 (0) |
| Alatasetal | 0 | - | - | - | ∞ | - | - | - | - | - | - |
| NICGAR | 25.4 | 0.28 (0.03) | 0.93 (0.02) | 3.65 (0.51) | ∞ | 0.89 (0.03) | 0.86 (0.03) | 0.98 (0.01) | 2.04 (0.04) | 0.82 (0.03) | 99.96 (0.1) |
| Fars | |||||||||||
| EARMGA | 86.40 | 0.23 (0.05) | 1 (0) | 1.01 (0) | ∞ | 0.03 (0.06) | 0.00 (0.00) | 0.01 (0.01) | 2 (0) | 0.67 (0.03) | 99.89 (0.25) |
| GAR | 202.60 | 0.86 (0.00) | 0.95 (0.00) | 1.05 (0.01) | ∞ | 0.34 (0.04) | 0.32 (0.04) | 0.35 (0.05) | 2.07 (0.03) | 0.31 (0.02) | 100 (0) |
| GENAR | 24.20 | 0.01 (0.00) | 0.87 (0.00) | 4.38 (0.01) | 10.01 | 0.84 (0.00) | 0.67 (0.01) | 0.83 (0.06) | 30.00 (0.00) | 0.44 (0.01) | 1.84 (0.15) |
| Alatasetal | 61.33 | 0.13 (0.20) | 1 (0) | 2.67 (2.75) | ∞ | 0.91 (0.16) | 0.20 (0.22) | 0.32 (0.09) | 8.42 (2.24) | 0.39 (0.13) | 39.68 (47.05) |
| NICGAR | 12.60 | 0.31 (0.05) | 0.99 (0.00) | 5.67 (0.58) | ∞ | 0.99 (0.01) | 0.98 (0.02) | 1 (0) | 2.02 (0.04) | 0.81 (0.04) | 100 (0) |
| House16 | |||||||||||
| EARMGA | 75.6 | 0.3 (0.1) | 1 (0) | 1 (0.01) | ∞ | 0.05 (0.08) | 0 (0.01) | 0 (0) | 2.2 (0.45) | 0.54 (0.09) | 99.95 (0.12) |
| GENAR | 30 | 0.45 (0) | 1 (0.01) | 1.01 (0) | 2.56 | 0.52 (0.02) | 0.02 (0) | 0.5 (0.02) | 17 (0) | 0.14 (0.01) | 86.94 (0.32) |
| GAR | 112.8 | 0.77 (0.01) | 0.9 (0) | 1.03 (0) | 1.33 | 0.2 (0) | 0.17 (0.01) | 0.49 (0.02) | 2.01 (0.01) | 0.28 (0.02) | 99.98 (0.01) |
| Alatasetal | 91 | 0.19 (0.01) | 0.99 (0.01) | 1.03 (0.02) | ∞ | 0.58 (0.16) | 0.03 (0.01) | 0.41 (0.15) | 8.75 (1.39) | 0.51 (0.03) | 98.24 (1.18) |
| NICGAR | 6 | 0.24 (0.04) | 0.92 (0) | 3.67 (0.71) | 15 | 0.88 (0.01) | 0.83 (0.01) | 0.98 (0) | 2 (0) | 0.86 (0.02) | 82.18 (13.91) |
| Ionosphere | |||||||||||
| EARMGA | 100 | 0.4 (0.1) | 1 (0) | 1 (0) | 1 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.59 (0.06) | 100 (0) |
| GENAR | 30 | 0.3 (0) | 0.99 (0) | 1.55 (0.01) | ∞ | 0.97 (0) | 0.5 (0) | 0.98 (0) | 34 (0) | 0.04 (0) | 34.99 (0.42) |
| GAR | 37.6 | 0.21 (0.01) | 0.92 (0.01) | 1.68 (0.04) | ∞ | 0.81 (0.02) | 0.44 (0.01) | 0.84 (0.01) | 2 (0) | 0.44 (0.04) | 81.26 (1.5) |
| Alatasetal | 96 | 0.79 (0.02) | 1 (0.01) | 1.01 (0.5) | ∞ | 0.43 (0.02) | 0.03 (0.02) | 0.49 (0.01) | 9.69 (0.02) | 0.05 (0.06) | 99.44 (1.4) |
| NICGAR | 24.8 | 0.2 (0.02) | 0.85 (0.01) | 3.77 (0.59) | ∞ | 0.78 (0.02) | 0.74 (0.02) | 0.96 (0) | 2.01 (0.02) | 0.88 (0.01) | 99.83 (0.15) |
| Letter | |||||||||||
| EARMGA | 100 | 0.36 (0.07) | 1 (0) | 1 (0) | ∞ | 0 (0.01) | 0 (0) | 0 (0.01) | 2 (0) | 0.6 (0.03) | 100 (0) |
| GENAR | 30 | 0.02 (0) | 0.27 (0.03) | 6.9 (0.66) | 1.92 | 0.24 (0.03) | 0.25 (0.03) | 0.82 (0.02) | 17 (0) | 0.63 (0.01) | 72.76 (1.23) |
| GAR | 13.4 | 0.6 (0.05) | 0.85 (0.01) | 1.15 (0.04) | 1.55 | 0.29 (0.06) | 0.25 (0.05) | 0.49 (0.06) | 2 (0) | 0.37 (0.12) | 99.95 (0.06) |
| Alatasetal | 35.33 | 0.21 (0.18) | 0.99 (0.01) | 4.93 (6.73) | ∞ | 0.76 (0.22) | 0.22 (0.28) | 0.36 (0.3) | 7 (2.66) | 0.42 (0.19) | 54.28 (46.04) |
| NICGAR | 18.8 | 0.15 (0.02) | 0.89 (0.02) | 5.94 (1.31) | ∞ | 0.84 (0.03) | 0.76 (0.04) | 0.96 (0.01) | 2.01 (0.02) | 0.87 (0.01) | 98.17 (1.62) |
| Magic | |||||||||||
| EARMGA | 96 | 0.33 (0.09) | 1 (0) | 1 (0) | ∞ | 0.01 (0.02) | 0 (0) | 0 (0.01) | 2 (0) | 0.58 (0.06) | 100 (0) |
| GENAR | 30 | 0.43 (0) | 0.81 (0) | 1.25 (0) | 1.85 | 0.46 (0) | 0.34 (0) | 0.66 (0) | 11 (0) | 0.04 (0) | 62.81 (0.76) |
| GAR | 64.2 | 0.67 (0.01) | 0.91 (0.01) | 1.11 (0.01) | 2.32 | 0.49 (0.02) | 0.35 (0.02) | 0.74 (0.02) | 2.11 (0.06) | 0.18 (0.02) | 97.47 (0.93) |
| Alatasetal | 11 | 0.47 (0.09) | 1 (0.01) | 1.26 (0.06) | 956.59 | 0.88 (0.02) | 0.34 (0.02) | 0.91 (0.02) | 4.73 (0.06) | 0.05 (0.06) | 89.9 (0.9) |
| NICGAR | 6.6 | 0.26 (0.01) | 0.94 (0.01) | 3 (0.06) | 15.3 | 0.91 (0.02) | 0.85 (0.01) | 0.99 (0.01) | 2 (0) | 0.85 (0.01) | 94.87 (4.93) |
| Movement Libras | |||||||||||
| EARMGA | 100 | 0.4 (0.03) | 1 (0) | 1 (0) | 1 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.62 (0.03) | 100 (0) |
| GENAR | 30 | 0.03 (0) | 0.91 (0.01) | 13.5 (0.1) | ∞ | 0.9 (0.01) | 0.87 (0.01) | 0.99 (0) | 91 (0) | 0.68 (0.01) | 55.23 (1.34) |
| GAR | 3 | 0.31 (0.2) | 0.89 (0.03) | 3.69 (1.93) | 6.52 | 0.83 (0.03) | 0.83 (0.03) | 0.99 (0.01) | 2 (0) | 0.29 (0.33) | 49.31 (23.1) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 27.8 | 0.27 (0.01) | 0.98 (0) | 3.64 (0.05) | ∞ | 0.97 (0.01) | 0.95 (0.01) | 1 (0) | 2 (0) | 0.86 (0) | 100 (0) |
| Optdigits | |||||||||||
| EARMGA | 97.8 | 0.41 (0.17) | 1 (0) | 1 (0.01) | ∞ | 0.03 (0.04) | 0 (0.01) | 0 (0.01) | 2 (0) | 0.59 (0.08) | 100 (0) |
| GENAR | 17 | 0.01 (0) | 1 (0) | 10.1 (0.01) | ∞ | 1 (0) | 0.91 (0) | 0.89 (0.04) | 63 (0) | 0.56 (0.01) | 2.11 (0.34) |
| GAR | 63.6 | 0.71 (0.02) | 0.97 (0) | 1.02 (0.02) | ∞ | 0.17 (0.03) | 0.04 (0.03) | 0.1 (0.08) | 2.01 (0.03) | 0.49 (0.01) | 100 (0) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 28.8 | 0.28 (0.02) | 0.87 (0.02) | 3.24 (0.4) | ∞ | 0.75 (0.05) | 0.71 (0.01) | 0.95 (0.01) | 2.05 (0.04) | 0.82 (0.02) | 100 (0) |
| Penbased | |||||||||||
| EARMGA | 100 | 0.42 (0.12) | 1 (0) | 1 (0) | 1 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.58 (0.06) | 100 (0) |
| GENAR | 30 | 0.05 (0) | 0.96 (0.01) | 9.56 (0.13) | ∞ | 0.96 (0.01) | 0.91 (0.01) | 1 (0) | 17 (0) | 0.62 (0.02) | 46.29 (1.2) |
| GAR | 2 | 0.72 (0) | 0.87 (0) | 1.04 (0) | 1.22 | 0.18 (0) | 0.18 (0) | 0.48 (0) | 2 (0) | 0 (0) | 94.95 (0) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 15 | 0.18 (0.02) | 0.89 (0.01) | 3.64 (0.77) | ∞ | 0.83 (0.02) | 0.73 (0.03) | 0.95 (0.01) | 2.06 (0.1) | 0.88 (0.02) | 98.64 (0.92) |
| Pollution | |||||||||||
| EARMGA | 100 | 0.21 (0.09) | 1 (0) | 1.08 (0.06) | ∞ | 0.16 (0.12) | 0.03 (0.02) | 0.14 (0.1) | 2 (0) | 0.7 (0.05) | 100 (0) |
| GENAR | 30 | 0.23 (0.01) | 1 (0) | 1.23 (0.01) | ∞ | 0.99 (0.01) | 0.24 (0.01) | 0.98 (0.02) | 16 (0) | 0.15 (0.02) | 47.33 (5.22) |
| GAR | 61.4 | 0.67 (0.03) | 0.92 (0.01) | 1.17 (0.02) | ∞ | 0.56 (0.05) | 0.46 (0.05) | 0.78 (0.05) | 2 (0) | 0.19 (0.02) | 100 (0) |
| Alatasetal | 15.4 | 0.59 (0.52) | 1 (0) | 6.86 (8.43) | ∞ | 0.43 (0.52) | 0.39 (0.51) | 0.43 (0.52) | 3.32 (0.94) | 0.01 (0.01) | 59.67 (52.98) |
| NICGAR | 57.8 | 0.08 (0.01) | 0.98 (0.01) | 8.35 (1.33) | ∞ | 0.97 (0.01) | 0.84 (0.02) | 1 (0) | 2.07 (0.05) | 0.93 (0.01) | 100 (0) |
| Quake | |||||||||||
| EARMGA | 100 | 0.3 (0.04) | 1 (0) | 1 (0) | 1 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.45 (0.03) | 100 (0) |
| GENAR | 30 | 0.55 (0) | 0.95 (0) | 1.01 (0) | 1.09 | 0.09 (0.01) | 0.01 (0.01) | 0.1 (0) | 4 (0) | 0.07 (0.01) | 81.89 (0.06) |
| GAR | 1 | 0.45 (0.02) | 0.85 (0.02) | 0.99 (0) | 0.91 | -0.02 (0) | -0.03 (0.01) | -0.13 (0.07) | 2 (0) | 0 (0) | 53.26 (1.13) |
| Alatasetal | 4.25 | 0.67 (0.18) | 1 (0.01) | 1.01 (0.01) | ∞ | 0.1 (0.11) | 0 (0.01) | 0 (0.08) | 2.08 (0.17) | 0.2 (0.19) | 98.06 (3.39) |
| NICGAR | 3.6 | 0.28 (0.05) | 0.9 (0.01) | 2.51 (0.8) | 11.4 | 0.81 (0.02) | 0.72 (0.02) | 0.95 (0.01) | 2 (0) | 0.75 (0.03) | 83.15 (1.7) |
| Satimage | |||||||||||
| EARMGA | 88.8 | 0.38 (0.1) | 1 (0) | 1 (0.01) | ∞ | 0.01 (0.02) | 0 (0.01) | 0 (0) | 2 (0) | 0.56 (0.03) | 100 (0) |
| GENAR | 30 | 0.22 (0) | 0.31 (0.01) | 1.42 (0.09) | ∞ | 0.1 (0.01) | 0.3 (0.01) | 0.97 (0.01) | 37 (0) | 0.25 (0.03) | 99.98 (0.03) |
| GAR | 206.6 | 0.91 (0.01) | 0.97 (0) | 1.04 (0.01) | 2.22 | 0.39 (0.02) | 0.37 (0.03) | 0.76 (0.05) | 2.1 (0.05) | 0.42 (0.01) | 100 (0) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 24 | 0.24 (0.01) | 0.92 (0.01) | 3.83 (0.31) | 12.27 | 0.88 (0.01) | 0.85 (0.01) | 0.99 (0) | 2 (0) | 0.82 (0.01) | 93.94 (2.29) |
| Segment | |||||||||||
| EARMGA | 99.4 | 0.39 (0.22) | 1 (0) | 1.05 (0.04) | ∞ | 0.07 (0.03) | 0.03 (0.01) | 0.06 (0.03) | 2 (0) | 0.58 (0.11) | 100 (0) |
| GENAR | 30 | 0.08 (0) | 0.77 (0.03) | 5.37 (0.19) | ∞ | 0.73 (0.03) | 0.7 (0.03) | 0.93 (0.01) | 20 (0) | 0.53 (0.02) | 85.13 (2.93) |
| GAR | 20.6 | 0.4 (0.08) | 0.89 (0.01) | 2.16 (0.3) | 3.45 | 0.52 (0.08) | 0.4 (0.05) | 0.64 (0.09) | 2 (0) | 0.4 (0.08) | 97.18 (1.6) |
| Alatasetal | 63 | 0.49 (0.25) | 0.96 (0.05) | 1.04 (0.03) | ∞ | 0.32 (0.16) | 0.03 (0.06) | 0.25 (0.21) | 4.26 (0.54) | 0.36 (0.18) | 99.96 (0.06) |
| NICGAR | 12.6 | 0.22 (0.01) | 0.99 (0.01) | 4.91 (0.43) | ∞ | 0.98 (0.01) | 0.97 (0.01) | 1 (0) | 2 (0) | 0.88 (0.01) | 99.76 (0.26) |
| Sonar | |||||||||||
| EARMGA | 100 | 0.33 (0.08) | 1 (0) | 1.03 (0.05) | ∞ | 0.03 (0.03) | 0.01 (0.02) | 0.02 (0.02) | 2 (0) | 0.62 (0.04) | 100 (0) |
| GENAR | 30 | 0.04 (0) | 0.94 (0.01) | 1.81 (0.03) | ∞ | 0.87 (0.03) | 0.44 (0.02) | 0.88 (0.03) | 61 (0) | 0.47 (0.03) | 30.96 (1.06) |
| GAR | 7.2 | 0.38 (0.05) | 0.83 (0.01) | 1.3 (0.05) | 2.05 | 0.45 (0.03) | 0.29 (0.03) | 0.59 (0.04) | 2 (0) | 0.21 (0.17) | 69.62 (21.6) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 22.8 | 0.26 (0.03) | 0.89 (0.02) | 7.56 (5.59) | ∞ | 0.81 (0.02) | 0.77 (0.02) | 0.97 (0.01) | 2.01 (0.02) | 0.84 (0.02) | 100 (0) |
| Spambase | |||||||||||
| EARMGA | 61.2 | 0.25 (0.16) | 1 (0) | 1.01 (0) | ∞ | 0.35 (0.36) | 0.01 (0) | 0 (0) | 2 (0) | 0.63 (0.13) | 81.46 (41.45) |
| GENAR | 30 | 0.45 (0) | 0.62 (0) | 1.03 (0.01) | 1.05 | 0.05 (0.01) | 0.06 (0.01) | 0.12 (0.02) | 58 (0) | 0.06 (0) | 86.21 (0.24) |
| GAR | 10.6 | 0.55 (0) | 0.9 (0) | 1.09 (0.01) | 1.74 | 0.41 (0.02) | 0.19 (0.01) | 0.55 (0.03) | 2 (0) | 0.02 (0.01) | 60.59 (0) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 11.2 | 0.12 (0.02) | 0.95 (0.02) | 73.02 (28.16) | ∞ | 0.9 (0.03) | 0.87 (0.04) | 0.98 (0.01) | 2 (0) | 0.84 (0.05) | 76.75 (31.96) |
| Spectfheart | |||||||||||
| EARMGA | 100 | 0.36 (0.1) | 1 (0) | 1.01 (0.02) | ∞ | 0.02 (0.04) | 0.01 (0.01) | 0.02 (0.03) | 2 (0) | 0.6 (0.06) | 100 (0) |
| GENAR | 30 | 0.25 (0) | 0.68 (0.01) | 0.91 (0.01) | 0.68 | -0.13 (0.01) | -0.17 (0.01) | -0.45 (0.02) | 45 (0) | 0.14 (0) | 60.15 (0.42) |
| GAR | 33.4 | 0.7 (0.02) | 0.89 (0.01) | 1.09 (0.01) | 1.69 | 0.35 (0.03) | 0.32 (0.04) | 0.67 (0.05) | 2 (0) | 0.42 (0.05) | 99.85 (0.33) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 72.2 | 0.13 (0.01) | 0.98 (0) | 29.01 (4.73) | ∞ | 0.92 (0) | 0.92 (0.02) | 1 (0) | 2.03 (0.05) | 0.89 (0.02) | 100 (0) |
| Stock | |||||||||||
| EARMGA | 100 | 0.38 (0.09) | 1 (0) | 1.01 (0.01) | ∞ | 0.01 (0.01) | 0.01 (0.01) | 0.01 (0.01) | 2 (0) | 0.57 (0.05) | 100 (0) |
| GENAR | 30 | 0.3 (0.01) | 0.92 (0.01) | 1.64 (0.04) | ∞ | 0.81 (0.03) | 0.52 (0.02) | 0.88 (0.02) | 10 (0) | 0.24 (0.01) | 87.54 (1.44) |
| GAR | 2 | 0.52 (0.11) | 0.88 (0.05) | 1.52 (0.18) | 3.74 | 0.72 (0.04) | 0.72 (0.04) | 0.95 (0.02) | 2 (0) | 0 (0) | 65.9 (8.3) |
| Alatasetal | 7 | 0.11 (0.2) | 0.99 (0.01) | 76.82 (135.96) | ∞ | 0.89 (0.24) | 0.71 (0.37) | 0.75 (0.29) | 2.57 (0.93) | 0.06 (0.13) | 20.45 (43.84) |
| NICGAR | 9.2 | 0.28 (0.02) | 0.95 (0.01) | 3.01 (0.18) | ∞ | 0.93 (0.02) | 0.87 (0.01) | 0.99 (0) | 2.07 (0.07) | 0.86 (0.02) | 99.41 (0.23) |
| Stulong | |||||||||||
| EARMGA | 94 | 0.3 (0.06) | 1 (0) | 1.01 (0) | ∞ | 0.06 (0.07) | 0.01 (0) | 0 (0) | 2 (0) | 0.5 (0.05) | 100 (0) |
| GENAR | 30 | 0.89 (0) | 0.99 (0) | 1.01 (0) | 1.02 | 0.02 (0) | 0.01 (0) | 0.06 (0.01) | 5 (0) | 0.02 (0) | 95.12 (0.35) |
| GAR | 161.6 | 0.78 (0.02) | 0.93 (0.01) | 1.03 (0.01) | 1.62 | 0.3 (0.02) | 0.2 (0.01) | 0.61 (0.03) | 2.98 (0.06) | 0.04 (0.01) | 99.94 (0.03) |
| Alatasetal | 7.67 | 0.72 (0.17) | 1 (0.01) | 1.48 (0.47) | ∞ | 0.23 (0.16) | 0.08 (0.04) | 0.11 (0.41) | 2.96 (0.34) | 0.08 (0.08) | 99.25 (0.7) |
| NICGAR | 4.2 | 0.26 (0.07) | 0.79 (0.03) | 16.82 (19.9) | ∞ | 0.62 (0.08) | 0.64 (0.06) | 0.9 (0.03) | 2.04 (0.09) | 0.78 (0.06) | 84.99 (7.01) |
| Texture | |||||||||||
| EARMGA | 100 | 0.24 (0.05) | 1 (0) | 1.17 (0.33) | ∞ | 0.08 (0.1) | 0.03 (0.03) | 0.07 (0.1) | 2 (0) | 0.63 (0.06) | 100 (0) |
| GENAR | 30 | 0.09 (0) | 0.68 (0.02) | 7.49 (0.2) | ∞ | 0.65 (0.02) | 0.68 (0.02) | 0.99 (0) | 41 (0) | 0.48 (0.03) | 98.25 (0.17) |
| GAR | 43.8 | 0.71 (0) | 0.93 (0.01) | 1.24 (0.02) | ∞ | 0.69 (0.02) | 0.66 (0.02) | 0.93 (0.01) | 2.01 (0.03) | 0.38 (0.03) | 97.85 (0.8) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 17.4 | 0.27 (0.02) | 0.97 (0) | 3.72 (0.39) | ∞ | 0.95 (0) | 0.92 (0.01) | 1 (0) | 2 (0) | 0.82 (0.02) | 95.85 (4.66) |
| Thyroid | |||||||||||
| EARMGA | 88 | 0.57 (0.11) | 1 (0) | 1 (0.01) | ∞ | 0.01 (0.01) | 0 (0.01) | 0 (0.01) | 2 (0) | 0.48 (0.04) | 100 (0) |
| GENAR | 30 | 0.69 (0) | 0.93 (0) | 1.01 (0) | 1.05 | 0.05 (0.01) | 0.02 (0.01) | 0.07 (0.01) | 22 (0) | 0.04 (0) | 95.5 (0.57) |
| GAR | 191.2 | 0.82 (0.01) | 0.92 (0) | 1.02 (0) | 1.17 | 0.13 (0.01) | 0.13 (0.01) | 0.45 (0.02) | 2.01 (0.02) | 0.03 (0) | 99.91 (0.08) |
| Alatasetal | 86.5 | 0.26 (0.1) | 0.97 (0.03) | 1.02 (0.01) | ∞ | 0.58 (0.07) | 0.05 (0.02) | 0.22 (0.12) | 16.75 (3.22) | 0.43 (0.02) | 89.92 (5.28) |
| NICGAR | 6.2 | 0.23 (0.07) | 0.93 (0.02) | 14.29 (9.46) | ∞ | 0.8 (0.07) | 0.85 (0.02) | 0.95 (0.05) | 2.02 (0.05) | 0.79 (0.05) | 98.16 (2.3) |
| Vehicle | |||||||||||
| EARMGA | 100 | 0.32 (0.07) | 1 (0) | 1.08 (0.07) | ∞ | 0.11 (0.08) | 0.04 (0.04) | 0.09 (0.07) | 2 (0) | 0.61 (0.04) | 100 (0) |
| GENAR | 30 | 0.09 (0.01) | 0.67 (0.02) | 2.62 (0.06) | 2.6 | 0.56 (0.02) | 0.48 (0.02) | 0.76 (0.02) | 19 (0) | 0.42 (0.01) | 66.39 (1.9) |
| GAR | 26.4 | 0.67 (0.04) | 0.94 (0.01) | 1.25 (0.14) | ∞ | 0.42 (0.07) | 0.3 (0.09) | 0.74 (0.09) | 2.02 (0.05) | 0.21 (0.08) | 100 (0) |
| Alatasetal | 22.4 | 0.01 (0) | 1 (0) | 77.37 (120.38) | ∞ | 1 (0) | 0.79 (0.15) | 0.61 (0.26) | 4.27 (1.01) | 0.16 (0.22) | 0.24 (0.12) |
| NICGAR | 12.8 | 0.27 (0.03) | 0.97 (0.01) | 3.46 (0.26) | ∞ | 0.95 (0.02) | 0.92 (0.01) | 1 (0) | 2 (0) | 0.86 (0.02) | 99.98 (0.05) |
| Wdbc | |||||||||||
| EARMGA | 100 | 0.27 (0.12) | 1 (0) | 1.09 (0.16) | ∞ | 0.06 (0.09) | 0.03 (0.06) | 0.05 (0.08) | 2 (0) | 0.61 (0.09) | 100 (0) |
| GENAR | 30 | 0.44 (0.01) | 0.94 (0) | 1.53 (0.01) | 6.42 | 0.84 (0.01) | 0.6 (0) | 0.94 (0) | 31 (0) | 0.08 (0) | 72.03 (1.52) |
| GAR | 90.4 | 0.54 (0.01) | 0.86 (0) | 1.18 (0.02) | 2.35 | 0.35 (0.03) | 0.29 (0.03) | 0.45 (0.04) | 2 (0) | 0.09 (0.02) | 99.13 (1.23) |
| Alatasetal | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 13.6 | 0.27 (0.03) | 0.96 (0.02) | 3.49 (0.39) | ∞ | 0.93 (0.04) | 0.9 (0.03) | 0.99 (0.01) | 2 (0) | 0.84 (0.01) | 98.53 (2.1) |
| Wine | |||||||||||
| EARMGA | 100 | 0.4 (0.14) | 1 (0) | 1.01 (0.01) | ∞ | 0.03 (0.03) | 0.01 (0.01) | 0.02 (0.03) | 2 (0) | 0.58 (0.08) | 100 (0) |
| GENAR | 30 | 0.2 (0.01) | 1 (0) | 3.02 (0.02) | ∞ | 1 (0) | 0.83 (0.01) | 1 (0) | 14 (0) | 0.28 (0.03) | 66.41 (1.84) |
| GAR | 7.6 | 0.21 (0.01) | 0.98 (0.03) | 2.83 (0.16) | ∞ | 0.96 (0.04) | 0.79 (0.04) | 0.99 (0.02) | 2 (0) | 0.54 (0.09) | 45.29 (4.32) |
| Alatasetal | 27 | 0.27 (0.34) | 1 (0) | 40.9 (55.02) | ∞ | 0.81 (0.28) | 0.47 (0.48) | 0.78 (0.33) | 4.34 (1.34) | 0.1 (0.11) | 40.23 (53.29) |
| NICGAR | 6.6 | 0.28 (0.01) | 0.94 (0.01) | 2.79 (0.18) | ∞ | 0.9 (0.02) | 0.84 (0.02) | 0.99 (0.01) | 2 (0) | 0.86 (0) | 94.61 (0.85) |
| Vowel | |||||||||||
| EARMGA | 100 | 0.34 (0.11) | 1 (0) | 1 (0) | ∞ | 0 (0.01) | 0 (0) | 0 (0.01) | 2 (0) | 0.62 (0.06) | 100 (0) |
| GENAR | 30 | 0.02 (0) | 0.56 (0.02) | 6.06 (0.28) | ∞ | 0.51 (0.03) | 0.48 (0.03) | 0.86 (0.01) | 14 (0) | 0.7 (0) | 63.64 (1.78) |
| GAR | 1.67 | 0.68 (0.08) | 0.86 (0.03) | 1.04 (0.01) | 1.23 | 0.18 (0.08) | 0.17 (0.1) | 0.44 (0.21) | 2 (0) | 0 (0) | 86.74 (13.44) |
| Alatasetal | 91.8 | 0.13 (0.02) | 1 (0) | 72.54 (138.87) | ∞ | 0.76 (0.09) | 0.17 (0.14) | 0.62 (0.15) | 8.36 (0.79) | 0.56 (0.01) | 94.47 (4.49) |
| NICGAR | 8.4 | 0.27 (0.11) | 0.95 (0.05) | 5.81 (2.57) | ∞ | 0.92 (0.07) | 0.89 (0.08) | 0.95 (0.09) | 2.02 (0.05) | 0.85 (0.01) | 100 (0) |
| Algorithm | #R | AvSup (σ) | AvConf (σ) | AvLift (σ) | AvConv | AvCF (σ) | AvNetconf (σ) | AvYulesQ (σ) | AvAmp (σ) | AvDiv (σ) | %Trans (σ) |
| Balance | |||||||||||
| QAR-CIP-NSGAII | 132.20 | 0.04 (0.00) | 0.92 (0.00) | 6.37 (0.31) | ∞ | 0.88 (0.01) | 0.68 (0.01) | 0.93 (0.00) | 3.97 (0.03) | 0.83 (0.00) | 91.72 (1.23) |
| MOPNAR | 96.80 | 0.16 (0.01) | 0.83 (0.02) | 2.02 (0.09) | ∞ | 0.70 (0.02) | 0.43 (0.00) | 0.81 (0.01) | 2.99 (0.07) | 0.64 (0.01) | 100.00 (0.00) |
| NICGAR | 26.40 | 0.06 (0.02) | 0.92 (0.03) | 4.25 (0.44) | ∞ | 0.86 (0.04) | 0.69 (0.03) | 0.91 (0.02) | 3.81 (0.18) | 0.82 (0.02) | 89.96 (2.19) |
| Basketball | |||||||||||
| QAR-CIP-NSGAII | 200.60 | 0.03 (0.01) | 0.99 (0.01) | 82.09 (2.07) | ∞ | 0.98 (0.01) | 0.96 (0.01) | 1.00 (0.00) | 2.10 (0.02) | 0.95 (0.01) | 96.67 (1.36) |
| MOPNAR | 134.20 | 0.13 (0.01) | 0.97 (0.01) | 58.43 (4.03) | ∞ | 0.94 (0.01) | 0.84 (0.02) | 0.98 (0.01) | 2.27 (0.04) | 0.75 (0.02) | 100.00 (0.00) |
| NICGAR | 49.80 | 0.04 (0.01) | 0.99 (0.01) | 9.43 (2.53) | ∞ | 0.98 (0.02) | 0.80 (0.04) | 0.99 (0.01) | 2.15 (0.09) | 0.95 (0.01) | 91.67 (4.04) |
| Bolts | |||||||||||
| QAR-CIP-NSGAII | 174.00 | 0.08 (0.01) | 1.00 (0.00) | 30.19 (1.97) | ∞ | 1.00 (0.00) | 0.99 (0.00) | 1.00 (0.00) | 2.10 (0.07) | 0.91 (0.03) | 100.00 (0.00) |
| MOPNAR | 69.00 | 0.37 (0.12) | 1.00 (0.00) | 15.77 (5.69) | ∞ | 1.00 (0.00) | 0.92 (0.04) | 1.00 (0.00) | 2.30 (0.12) | 0.54 (0.15) | 100.00 (0.00) |
| NICGAR | 9.80 | 0.30 (0.04) | 0.98 (0.02) | 5.51 (1.66) | ∞ | 0.98 (0.02) | 0.93 (0.03) | 1.00 (0.00) | 2.00 (0.00) | 0.82 (0.02) | 100.00 (0.00) |
| Coil2000 | |||||||||||
| QAR-CIP-NSGAII | 58.60 | 0.26 (0.11) | 0.93 (0.04) | 127.77 (103.26) | ∞ | 0.93 (0.04) | 0.80 (0.13) | 0.86 (0.13) | 3.33 (0.63) | 0.72 (0.05) | 100.00 (0.00) |
| MOPNAR | 39.20 | 0.41 (0.04) | 0.92 (0.08) | 6.80 (3.63) | ∞ | 0.89 (0.08) | 0.71 (0.04) | 0.97 (0.01) | 3.15 (0.17) | 0.55 (0.07) | 100.00 (0.00) |
| NICGAR | 25.40 | 0.28 (0.03) | 0.93 (0.02) | 3.65 (0.51) | ∞ | 0.89 (0.03) | 0.86 (0.03) | 0.98 (0.01) | 2.04 (0.04) | 0.82 (0.03) | 99.96 (0.10) |
| Fars | |||||||||||
| QAR-CIP-NSGAII | 109.40 | 0.22 (0.02) | 0.95 (0.01) | 544.76 (220.80) | ∞ | 0.95 (0.02) | 0.75 (0.03) | 0.84 (0.04) | 3.33 (0.05) | 0.78 (0.02) | 100.00 (0.00) |
| MOPNAR | 58.00 | 0.34 (0.02) | 0.90 (0.05) | 8.00 (1.79) | ∞ | 0.89 (0.05) | 0.83 (0.05) | 1.00 (0.00) | 2.73 (0.12) | 0.60 (0.04) | 100.00 (0.00) |
| NICGAR | 12.60 | 0.31 (0.05) | 0.99 (0.00) | 5.67 (0.58) | ∞ | 0.99 (0.01) | 0.98 (0.02) | 1.00 (0.00) | 2.02 (0.04) | 0.81 (0.04) | 100.00 (0.00) |
| House16H | |||||||||||
| QAR-CIP-NSGAII | 288.20 | 0.18 (0.02) | 0.93 (0.00) | 2549.17 (529.64) | ∞ | 0.91 (0.01) | 0.71 (0.03) | 0.84 (0.02) | 2.98 (0.17) | 0.76 (0.01) | 99.81 (0.35) |
| MOPNAR | 135.40 | 0.31 (0.07) | 0.92 (0.05) | 9.65 (3.06) | ∞ | 0.89 (0.05) | 0.75 (0.03) | 0.99 (0.00) | 2.79 (0.21) | 0.49 (0.14) | 99.98 (0.02) |
| NICGAR | 6.00 | 0.24 (0.04) | 0.92 (0.00) | 3.67 (0.71) | 15.00 | 0.88 (0.01) | 0.83 (0.01) | 0.98 (0.00) | 2.00 (0.00) | 0.86 (0.02) | 82.18 (13.91) |
| Ionosphere | |||||||||||
| QAR-CIP-NSGAII | 125.20 | 0.10 (0.02) | 0.96 (0.01) | 143.56 (22.61) | ∞ | 0.94 (0.01) | 0.90 (0.02) | 0.99 (0.00) | 2.67 (0.17) | 0.87 (0.05) | 87.70 (4.00) |
| MOPNAR | 91.20 | 0.32 (0.04) | 0.94 (0.02) | 12.59 (3.57) | ∞ | 0.89 (0.02) | 0.72 (0.04) | 0.98 (0.01) | 2.98 (0.06) | 0.63 (0.05) | 99.72 (0.64) |
| NICGAR | 24.80 | 0.20 (0.02) | 0.85 (0.01) | 3.77 (0.59) | ∞ | 0.78 (0.02) | 0.74 (0.02) | 0.96 (0.00) | 2.01 (0.02) | 0.88 (0.01) | 99.83 (0.15) |
| Letter | |||||||||||
| QAR-CIP-NSGAII | 105.20 | 0.09 (0.01) | 0.89 (0.01) | 371.41 (84.60) | ∞ | 0.87 (0.01) | 0.71 (0.03) | 0.84 (0.05) | 3.50 (0.17) | 0.79 (0.04) | 97.94 (1.87) |
| MOPNAR | 75.60 | 0.28 (0.03) | 0.91 (0.03) | 7.63 (1.25) | ∞ | 0.87 (0.01) | 0.64 (0.06) | 0.98 (0.01) | 3.33 (0.23) | 0.39 (0.04) | 99.94 (0.10) |
| NICGAR | 18.80 | 0.15 (0.02) | 0.89 (0.02) | 5.94 (1.31) | ∞ | 0.84 (0.03) | 0.76 (0.04) | 0.96 (0.01) | 2.01 (0.02) | 0.87 (0.01) | 98.17 (1.62) |
| Magic | |||||||||||
| QAR-CIP-NSGAII | 210.80 | 0.22 (0.04) | 0.95 (0.01) | 4464.87 (614.11) | ∞ | 0.93 (0.00) | 0.56 (0.04) | 0.71 (0.05) | 2.35 (0.05) | 0.74 (0.03) | 99.95 (0.03) |
| MOPNAR | 115.20 | 0.37 (0.05) | 0.91 (0.04) | 8.72 (1.44) | ∞ | 0.89 (0.03) | 0.70 (0.03) | 0.99 (0.01) | 2.58 (0.11) | 0.52 (0.05) | 99.98 (0.03) |
| NICGAR | 6.60 | 0.26 (0.01) | 0.94 (0.01) | 3.00 (0.06) | 15.30 | 0.91 (0.02) | 0.85 (0.01) | 0.99 (0.01) | 2.00 (0.00) | 0.85 (0.01) | 94.87 (4.93) |
| Movement Libras | |||||||||||
| QAR-CIP-NSGAII | 57.80 | 0.05 (0.01) | 0.96 (0.02) | 154.26 (26.59) | ∞ | 0.96 (0.02) | 0.94 (0.02) | 1.00 (0.00) | 2.37 (0.15) | 0.90 (0.03) | 55.28 (14.04) |
| MOPNAR | 84.20 | 0.26 (0.08) | 0.98 (0.01) | 24.01 (17.48) | ∞ | 0.97 (0.01) | 0.91 (0.02) | 1.00 (0.00) | 2.62 (0.16) | 0.62 (0.10) | 99.34 (1.20) |
| NICGAR | 27.80 | 0.27 (0.01) | 0.98 (0.01) | 3.64 (0.05) | ∞ | 0.97 (0.01) | 0.95 (0.01) | 1.00 (0.00) | 2.00 (0.00) | 0.86 (0.00) | 100.00 (0.00) |
| Optdigits | |||||||||||
| QAR-CIP-NSGAII | 94.60 | 0.19 (0.03) | 0.87 (0.03) | 40.21 (21.97) | ∞ | 0.84 (0.04) | 0.58 (0.05) | 0.85 (0.02) | 3.46 (0.21) | 0.69 (0.04) | 100.00 (0.00) |
| MOPNAR | 77.00 | 0.26 (0.07) | 0.85 (0.05) | 7.09 (2.53) | ∞ | 0.81 (0.05) | 0.60 (0.05) | 0.97 (0.01) | 3.45 (0.40) | 0.59 (0.02) | 100.00 (0.00) |
| NICGAR | 28.80 | 0.28 (0.02) | 0.87 (0.02) | 3.24 (0.40) | ∞ | 0.75 (0.05) | 0.71 (0.01) | 0.95 (0.01) | 2.05 (0.04) | 0.82 (0.02) | 100.00 (0.00) |
| Penbased | |||||||||||
| QAR-CIP-NSGAII | 108.40 | 0.07 (0.01) | 0.89 (0.03) | 243.31 (157.03) | ∞ | 0.87 (0.03) | 0.68 (0.08) | 0.82 (0.08) | 3.23 (0.26) | 0.85 (0.02) | 95.58 (2.79) |
| MOPNAR | 107.20 | 0.30 (0.07) | 0.92 (0.01) | 6.82 (1.69) | ∞ | 0.89 (0.02) | 0.73 (0.02) | 0.99 (0.00) | 2.99 (0.14) | 0.59 (0.01) | 99.98 (0.03) |
| NICGAR | 15.00 | 0.18 (0.02) | 0.89 (0.01) | 3.64 (0.77) | ∞ | 0.83 (0.02) | 0.73 (0.03) | 0.95 (0.01) | 2.06 (0.10) | 0.88 (0.02) | 98.64 (0.92) |
| Pollution | |||||||||||
| QAR-CIP-NSGAII | 249.40 | 0.05 (0.01) | 1.00 (0.01) | 51.75 (0.80) | ∞ | 0.99 (0.00) | 0.97 (0.01) | 1.00 (0.00) | 2.09 (0.02) | 0.95 (0.00) | 100.00 (0.00) |
| MOPNAR | 79.40 | 0.19 (0.03) | 0.99 (0.01) | 38.06 (3.84) | ∞ | 0.98 (0.01) | 0.90 (0.03) | 1.00 (0.00) | 2.25 (0.07) | 0.65 (0.11) | 98.00 (2.74) |
| NICGAR | 57.80 | 0.08 (0.01) | 0.98 (0.01) | 8.35 (1.33) | ∞ | 0.97 (0.01) | 0.84 (0.02) | 1.00 (0.00) | 2.07 (0.05) | 0.93 (0.01) | 100.00 (0.00) |
| Quake | |||||||||||
| QAR-CIP-NSGAII | 137.20 | 0.08 (0.01) | 0.93 (0.01) | 450.67 (46.72) | ∞ | 0.89 (0.01) | 0.62 (0.03) | 0.77 (0.02) | 2.32 (0.06) | 0.83 (0.02) | 71.60 (2.25) |
| MOPNAR | 49.20 | 0.32 (0.05) | 0.91 (0.04) | 8.70 (2.47) | ∞ | 0.86 (0.05) | 0.57 (0.03) | 0.95 (0.02) | 2.32 (0.09) | 0.52 (0.04) | 100.00 (0.00) |
| NICGAR | 3.60 | 0.28 (0.05) | 0.90 (0.01) | 2.51 (0.80) | 11.40 | 0.81 (0.02) | 0.72 (0.02) | 0.95 (0.01) | 2.00 (0.00) | 0.75 (0.03) | 83.15 (1.70) |
| Satimage | |||||||||||
| QAR-CIP-NSGAII | 299.80 | 0.29 (0.02) | 0.93 (0.01) | 34.04 (34.62) | ∞ | 0.90 (0.01) | 0.78 (0.02) | 0.96 (0.02) | 5.26 (0.36) | 0.69 (0.02) | 100.00 (0.00) |
| MOPNAR | 172.80 | 0.32 (0.04) | 0.95 (0.01) | 7.55 (0.61) | ∞ | 0.93 (0.01) | 0.80 (0.02) | 1.00 (0.01) | 3.74 (0.24) | 0.63 (0.05) | 100.00 (0.00) |
| NICGAR | 24.00 | 0.24 (0.01) | 0.92 (0.01) | 3.83 (0.31) | 12.27 | 0.88 (0.01) | 0.85 (0.01) | 0.99 (0.00) | 2.00 (0.00) | 0.82 (0.01) | 93.94 (2.29) |
| Segment | |||||||||||
| QAR-CIP-NSGAII | 176.60 | 0.18 (0.03) | 1.00 (0.00) | 451.22 (40.06) | ∞ | 0.99 (0.00) | 0.71 (0.02) | 0.79 (0.02) | 2.46 (0.07) | 0.83 (0.01) | 100.00 (0.00) |
| MOPNAR | 123.60 | 0.30 (0.01) | 0.99 (0.01) | 17.70 (2.35) | ∞ | 0.98 (0.01) | 0.89 (0.05) | 1.00 (0.00) | 2.66 (0.13) | 0.65 (0.04) | 99.99 (0.02) |
| NICGAR | 12.60 | 0.22 (0.01) | 0.99 (0.01) | 4.91 (0.43) | ∞ | 0.98 (0.01) | 0.97 (0.01) | 1.00 (0.00) | 2.00 (0.00) | 0.88 (0.01) | 99.76 (0.26) |
| Sonar | |||||||||||
| QAR-CIP-NSGAII | 123.00 | 0.06 (0.02) | 0.95 (0.02) | 128.39 (15.96) | ∞ | 0.92 (0.03) | 0.87 (0.04) | 0.97 (0.01) | 2.38 (0.10) | 0.91 (0.04) | 80.20 (12.03) |
| MOPNAR | 71.00 | 0.32 (0.05) | 0.93 (0.02) | 13.38 (7.66) | ∞ | 0.89 (0.03) | 0.71 (0.06) | 0.98 (0.01) | 2.74 (0.14) | 0.64 (0.04) | 99.14 (1.68) |
| NICGAR | 22.80 | 0.26 (0.03) | 0.89 (0.02) | 7.56 (5.59) | ∞ | 0.81 (0.02) | 0.77 (0.02) | 0.97 (0.01) | 2.01 (0.02) | 0.84 (0.02) | 100.00 (0.00) |
| Spambase | |||||||||||
| QAR-CIP-NSGAII | 178.60 | 0.29 (0.03) | 0.92 (0.02) | 154.34 (19.12) | ∞ | 0.86 (0.03) | 0.62 (0.01) | 0.91 (0.02) | 4.50 (0.26) | 0.64 (0.03) | 100.00 (0.00) |
| MOPNAR | 83.40 | 0.37 (0.08) | 0.86 (0.07) | 6.79 (1.09) | ∞ | 0.79 (0.07) | 0.58 (0.07) | 0.94 (0.02) | 4.17 (0.84) | 0.58 (0.03) | 100.00 (0.00) |
| NICGAR | 11.20 | 0.12 (0.02) | 0.95 (0.02) | 73.02 (28.16) | ∞ | 0.90 (0.03) | 0.87 (0.04) | 0.98 (0.01) | 2.00 (0.00) | 0.84 (0.05) | 76.75 (31.96) |
| Spectfheart | |||||||||||
| QAR-CIP-NSGAII | 109.00 | 0.17 (0.02) | 0.90 (0.02) | 55.74 (9.50) | ∞ | 0.84 (0.03) | 0.70 (0.04) | 0.94 (0.01) | 3.04 (0.19) | 0.79 (0.02) | 96.11 (1.53) |
| MOPNAR | 55.80 | 0.42 (0.02) | 0.92 (0.03) | 12.27 (5.12) | ∞ | 0.86 (0.03) | 0.64 (0.05) | 0.96 (0.02) | 2.70 (0.08) | 0.60 (0.05) | 99.70 (0.67) |
| NICGAR | 72.20 | 0.13 (0.01) | 0.98 (0.00) | 29.01 (4.73) | ∞ | 0.92 (0.00) | 0.92 (0.02) | 1.00 (0.00) | 2.03 (0.05) | 0.89 (0.02) | 100.00 (0.00) |
| Stock | |||||||||||
| QAR-CIP-NSGAII | 107.60 | 0.08 (0.01) | 0.94 (0.01) | 91.91 (43.28) | ∞ | 0.93 (0.01) | 0.90 (0.01) | 1.00 (0.00) | 2.97 (0.07) | 0.83 (0.04) | 73.75 (6.75) |
| MOPNAR | 105.20 | 0.26 (0.02) | 0.93 (0.01) | 12.51 (1.32) | ∞ | 0.92 (0.01) | 0.83 (0.02) | 1.00 (0.00) | 2.95 (0.14) | 0.59 (0.03) | 100.00 (0.00) |
| NICGAR | 9.20 | 0.28 (0.02) | 0.95 (0.01) | 3.01 (0.18) | ∞ | 0.93 (0.02) | 0.87 (0.01) | 0.99 (0.00) | 2.07 (0.07) | 0.86 (0.02) | 99.41 (0.23) |
| Stulong | |||||||||||
| QAR-CIP-NSGAII | 153.80 | 0.19 (0.03) | 0.82 (0.01) | 39.32 (5.96) | ∞ | 0.74 (0.01) | 0.58 (0.02) | 0.92 (0.01) | 2.90 (0.05) | 0.60 (0.02) | 99.94 (0.09) |
| MOPNAR | 89.40 | 0.31 (0.03) | 0.84 (0.02) | 4.33 (0.38) | ∞ | 0.76 (0.02) | 0.52 (0.02) | 0.93 (0.01) | 2.69 (0.14) | 0.50 (0.03) | 100.00 (0.00) |
| NICGAR | 4.20 | 0.26 (0.07) | 0.79 (0.03) | 16.82 (19.90) | ∞ | 0.62 (0.08) | 0.64 (0.06) | 0.90 (0.03) | 2.04 (0.09) | 0.78 (0.06) | 84.99 (7.01) |
| Texture | |||||||||||
| QAR-CIP-NSGAII | 151.00 | 0.14 (0.03) | 0.98 (0.01) | 785.39 (104.79) | ∞ | 0.97 (0.01) | 0.79 (0.02) | 0.84 (0.02) | 2.66 (0.16) | 0.78 (0.03) | 99.87 (0.14) |
| MOPNAR | 117.60 | 0.29 (0.06) | 0.97 (0.02) | 14.26 (4.86) | ∞ | 0.96 (0.02) | 0.90 (0.04) | 1.00 (0.00) | 2.83 (0.34) | 0.62 (0.08) | 100.00 (0.00) |
| NICGAR | 17.40 | 0.27 (0.02) | 0.97 (0.00) | 3.72 (0.39) | ∞ | 0.95 (0.00) | 0.92 (0.01) | 1.00 (0.00) | 2.00 (0.00) | 0.82 (0.02) | 95.85 (4.66) |
| Thyroid | |||||||||||
| QAR-CIP-NSGAII | 216.60 | 0.29 (0.03) | 0.93 (0.01) | 163.67 (40.48) | ∞ | 0.88 (0.01) | 0.63 (0.02) | 0.92 (0.01) | 3.30 (0.15) | 0.57 (0.03) | 100.00 (0.00) |
| MOPNAR | 75.60 | 0.37 (0.07) | 0.92 (0.03) | 11.43 (0.97) | ∞ | 0.84 (0.02) | 0.59 (0.04) | 0.94 (0.01) | 3.40 (0.56) | 0.43 (0.06) | 100.00 (0.01) |
| NICGAR | 6.20 | 0.23 (0.07) | 0.93 (0.02) | 14.29 (9.46) | ∞ | 0.80 (0.07) | 0.85 (0.02) | 0.95 (0.05) | 2.02 (0.05) | 0.79 (0.05) | 98.16 (2.30) |
| Wdbc | |||||||||||
| QAR-CIP-NSGAII | 126.80 | 0.17 (0.02) | 0.99 (0.00) | 220.99 (35.03) | ∞ | 0.98 (0.00) | 0.96 (0.01) | 1.00 (0.00) | 2.26 (0.07) | 0.85 (0.03) | 98.57 (1.24) |
| MOPNAR | 85.00 | 0.33 (0.02) | 0.98 (0.00) | 15.45 (3.37) | ∞ | 0.97 (0.00) | 0.91 (0.02) | 1.00 (0.00) | 2.56 (0.06) | 0.58 (0.12) | 99.58 (0.51) |
| NICGAR | 13.60 | 0.27 (0.03) | 0.96 (0.02) | 3.49 (0.39) | ∞ | 0.93 (0.04) | 0.90 (0.03) | 0.99 (0.01) | 2.00 (0.00) | 0.84 (0.01) | 98.53 (2.10) |
| Vehicle | |||||||||||
| QAR-CIP-NSGAII | 131.80 | 0.16 (0.01) | 0.99 (0.00) | 107.11 (23.20) | ∞ | 0.98 (0.01) | 0.96 (0.01) | 1.00 (0.00) | 2.35 (0.04) | 0.83 (0.01) | 100.00 (0.00) |
| MOPNAR | 107.40 | 0.27 (0.03) | 0.98 (0.01) | 15.39 (2.89) | ∞ | 0.98 (0.01) | 0.92 (0.02) | 1.00 (0.00) | 2.55 (0.09) | 0.64 (0.05) | 100.00 (0.00) |
| NICGAR | 12.80 | 0.27 (0.03) | 0.97 (0.01) | 3.46 (0.26) | ∞ | 0.95 (0.02) | 0.92 (0.01) | 1.00 (0.00) | 2.00 (0.00) | 0.86 (0.02) | 99.98 (0.05) |
| Wine | |||||||||||
| QAR-CIP-NSGAII | 122.20 | 0.04 (0.00) | 0.97 (0.01) | 118.50 (4.75) | ∞ | 0.97 (0.01) | 0.95 (0.01) | 1.00 (0.00) | 2.31 (0.09) | 0.92 (0.02) | 83.94 (9.20) |
| MOPNAR | 71.60 | 0.26 (0.03) | 0.95 (0.02) | 19.42 (9.74) | ∞ | 0.93 (0.03) | 0.81 (0.05) | 1.00 (0.00) | 2.83 (0.11) | 0.59 (0.03) | 100.00 (0.00) |
| NICGAR | 6.60 | 0.28 (0.01) | 0.94 (0.01) | 2.79 (0.18) | ∞ | 0.90 (0.02) | 0.84 (0.02) | 0.99 (0.01) | 2.00 (0.00) | 0.86 (0.00) | 94.61 (0.85) |
| Vowel | |||||||||||
| QAR-CIP-NSGAII | 84.40 | 0.03 (0.00) | 0.95 (0.02) | 540.52 (58.59) | ∞ | 0.95 (0.02) | 0.92 (0.02) | 0.99 (0.01) | 2.48 (0.12) | 0.94 (0.01) | 84.19 (9.37) |
| MOPNAR | 50.20 | 0.18 (0.03) | 0.91 (0.04) | 16.29 (2.97) | ∞ | 0.90 (0.04) | 0.75 (0.05) | 0.99 (0.01) | 3.21 (0.20) | 0.58 (0.05) | 99.72 (0.63) |
| NICGAR | 8.40 | 0.27 (0.11) | 0.95 (0.05) | 5.81 (2.57) | ∞ | 0.92 (0.07) | 0.89 (0.08) | 0.95 (0.09) | 2.02 (0.05) | 0.85 (0.01) | 100.00 (0.00) |
Results obtained in the comparison with classical algorithms
In this section, we present the results obtained by our algorithm against two classical algorithms (Apriori and Eclat). Table 5 shows the average results obtained of 5 runs for each dataset. Notice that we only show the results obtained by the Apriori and Eclat algorithms over 15 datasets due to the fact that they present scalability problems and cannot run in all datasets.
Finally, this performance results are available for download in xls, pdf and tex files by clicking the links ![]() ,
, ![]() and
and ![]() , respectively.
, respectively.
| Algorithm | #R | AvSup (σ) | AvConf (σ) | AvLift (σ) | AvConv | AvCF (σ) | AvNetconf (σ) | AvYulesQ (σ) | AvAmp (σ) | AvDiv (σ) | %Trans (σ) |
| Balance | |||||||||||
| Apriori | 2 | 0.14 (0) | 0.86 (0) | 6.25 (0) | 5.77 | 0.83 (0) | 0.86 (0) | 1 (0) | 3 (0) | 1 (0) | 27.2 (0) |
| Eclat | 2 | 0.14 (0) | 0.86 (0) | 6.25 (0) | 5.77 | 0.83 (0) | 0.86 (0) | 1 (0) | 3 (0) | 1 (0) | 27.2 (0) |
| NICGAR | 26.4 | 0.06 (0.02) | 0.92 (0.03) | 4.25 (0.44) | ∞ | 0.86 (0.04) | 0.69 (0.03) | 0.91 (0.02) | 3.81 (0.18) | 0.82 (0.02) | 89.96 (2.19) |
| Basketball | |||||||||||
| Apriori | 4 | 0.15 (0) | 0.87 (0) | 4.88 (0) | 6.65 | 0.84 (0) | 0.81 (0) | 0.99 (0) | 2.75 (0) | 0.5 (0) | 33.34 (0) |
| Eclat | 4 | 0.15 (0) | 0.87 (0) | 4.88 (0) | 6.65 | 0.84 (0) | 0.81 (0) | 0.99 (0) | 2.75 (0) | 0.5 (0) | 33.34 (0) |
| NICGAR | 49.8 | 0.04 (0.01) | 0.99 (0.01) | 9.43 (2.53) | ∞ | 0.98 (0.02) | 0.8 (0.04) | 0.99 (0.01) | 2.15 (0.09) | 0.95 (0.01) | 91.67 (4.04) |
| Bolts | |||||||||||
| Apriori | 1246 | 0.15 (0) | 0.99 (0) | 7.16 (0) | ∞ | 0.98 (0) | 0.96 (0) | 1 (0) | 4.36 (0) | 0.45 (0) | 97.5 (0) |
| Eclat | 1246 | 0.15 (0) | 0.99 (0) | 7.16 (0) | ∞ | 0.98 (0) | 0.96 (0) | 1 (0) | 4.36 (0) | 0.45 (0) | 97.5 (0) |
| NICGAR | 9.8 | 0.3 (0.04) | 0.98 (0.02) | 5.51 (1.66) | ∞ | 0.98 (0.02) | 0.93 (0.03) | 1 (0) | 2 (0) | 0.82 (0.02) | 100 (0) |
| Coil2000 | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 25.4 | 0.28 (0.03) | 0.93 (0.02) | 3.65 (0.51) | ∞ | 0.89 (0.03) | 0.86 (0.03) | 0.98 (0.01) | 2.04 (0.04) | 0.82 (0.03) | 99.96 (0.1) |
| Fars | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 12.6 | 0.31 (0.05) | 0.99 (0.00) | 5.67 (0.58) | ∞ | 0.99 (0.01) | 0.98 (0.02) | 1 (0) | 2.02 (0.04) | 0.81 (0.04) | 100 (0) |
| House16H | |||||||||||
| Apriori | 1749917 | 0.22 (0) | 0.97 (0) | 2.19 (0) | ∞ | 0.83 (0) | 0.45 (0) | 0.76 (0) | 8.65 (0) | 0.28 (0) | 100 (0) |
| Eclat | 1749917 | 0.22 (0) | 0.97 (0) | 2.19 (0) | ∞ | 0.83 (0) | 0.45 (0) | 0.76 (0) | 8.65 (0) | 0.37 (0) | 100 (0) |
| NICGAR | 6 | 0.24 (0.04) | 0.92 (0) | 3.67 (0.71) | 15 | 0.88 (0.01) | 0.83 (0.01) | 0.98 (0) | 2 (0) | 0.86 (0.02) | 82.18 (13.91) |
| Inosphere | |||||||||||
| Apriori | 211770656 | 0.12 (0) | 0.98 (0) | 4.4 (0) | ∞ | 0.97 (0) | 0.72 (0) | 1 (0) | 10.79 (0) | 0.03 (0) | 100 (0) |
| Eclat | 211770656 | 0.12 (0) | 0.98 (0) | 4.4 (0) | ∞ | 0.97 (0) | 0.72 (0) | 1 (0) | 10.79 (0) | 0.28 (0) | 100 (0) |
| NICGAR | 24.8 | 0.2 (0.02) | 0.85 (0.01) | 3.77 (0.59) | ∞ | 0.78 (0.02) | 0.74 (0.02) | 0.96 (0) | 2.01 (0.02) | 0.88 (0.01) | 99.83 (0.15) |
| Letter | |||||||||||
| Apriori | 3916 | 0.14 (0) | 0.88 (0) | 4.66 (0) | ∞ | 0.81 (0) | 0.73 (0) | 0.93 (0) | 4.55 (0) | 0.49 (0) | 99.49 (0) |
| Eclat | 3916 | 0.14 (0) | 0.88 (0) | 4.66 (0) | ∞ | 0.81 (0) | 0.73 (0) | 0.93 (0) | 4.55 (0) | 0.49 (0) | 99.49 (0) |
| NICGAR | 18.8 | 0.15 (0.02) | 0.89 (0.02) | 5.94 (1.31) | ∞ | 0.84 (0.03) | 0.76 (0.04) | 0.96 (0.01) | 2.01 (0.02) | 0.87 (0.01) | 98.17 (1.62) |
| Magic | |||||||||||
| Apriori | 9785 | 0.19 (0) | 0.96 (0) | 2.73 (0) | ∞ | 0.87 (0) | 0.52 (0) | 0.9 (0) | 5.53 (0) | 0.4 (0) | 99.96 (0) |
| Eclat | 9785 | 0.19 (0) | 0.96 (0) | 2.73 (0) | ∞ | 0.87 (0) | 0.52 (0) | 0.9 (0) | 5.53 (0) | 0.4 (0) | 99.96 (0) |
| NICGAR | 6.6 | 0.26 (0.01) | 0.94 (0.01) | 3 (0.06) | 15.3 | 0.91 (0.02) | 0.85 (0.01) | 0.99 (0.01) | 2 (0) | 0.85 (0.01) | 94.87 (4.93) |
| Movement Libras | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 27.8 | 0.27 (0.01) | 0.98 (0) | 3.64 (0.05) | ∞ | 0.97 (0.01) | 0.95 (0.01) | 1 (0) | 2 (0) | 0.86 (0) | 100 (0) |
| Optdigits | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 28.8 | 0.28 (0.02) | 0.87 (0.02) | 3.24 (0.4) | ∞ | 0.75 (0.05) | 0.71 (0.01) | 0.95 (0.01) | 2.05 (0.04) | 0.82 (0.02) | 100 (0) |
| Penbased | |||||||||||
| Apriori | 1918 | 0.14 (0) | 0.92 (0) | 4.23 (0) | ∞ | 0.82 (0) | 0.63 (0) | 0.87 (0) | 4.07 (0) | 0.5 (0) | 99.5 (0) |
| Eclat | 1918 | 0.14 (0) | 0.92 (0) | 4.23 (0) | ∞ | 0.82 (0) | 0.63 (0) | 0.87 (0) | 4.07 (0) | 0.5 (0) | 99.5 (0) |
| NICGAR | 15 | 0.18 (0.02) | 0.89 (0.01) | 3.64 (0.77) | ∞ | 0.83 (0.02) | 0.73 (0.03) | 0.95 (0.01) | 2.06 (0.1) | 0.88 (0.02) | 98.64 (0.92) |
| Pollution | |||||||||||
| Apriori | 41510 | 0.13 (0) | 0.95 (0) | 5.84 (0) | ∞ | 0.93 (0) | 0.86 (0) | 0.98 (0) | 5.88 (0) | 0.38 (0) | 100 (0) |
| Eclat | 41510 | 0.13 (0) | 0.95 (0) | 5.84 (0) | ∞ | 0.93 (0) | 0.86 (0) | 0.98 (0) | 5.88 (0) | 0.38 (0) | 100 (0) |
| NICGAR | 57.8 | 0.08 (0.01) | 0.98 (0.01) | 8.35 (1.33) | ∞ | 0.97 (0.01) | 0.84 (0.02) | 1 (0) | 2.07 (0.05) | 0.93 (0.01) | 100 (0) |
| Quake | |||||||||||
| Apriori | 18 | 0.25 (0) | 0.91 (0) | 1 (0) | 1.15 | 0.11 (0) | -0.01 (0) | 0.02 (0) | 2.56 (0) | 0.45 (0) | 90.55 (0) |
| Eclat | 18 | 0.25 (0) | 0.91 (0) | 1 (0) | 1.15 | 0.11 (0) | -0.01 (0) | 0.02 (0) | 2.56 (0) | 0.45 (0) | 90.55 (0) |
| NICGAR | 3.6 | 0.28 (0.05) | 0.9 (0.01) | 2.51 (0.8) | 11.4 | 0.81 (0.02) | 0.72 (0.02) | 0.95 (0.01) | 2 (0) | 0.75 (0.03) | 83.15 (1.7) |
| Satimage | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 24 | 0.24 (0.01) | 0.92 (0.01) | 3.83 (0.31) | 12.27 | 0.88 (0.01) | 0.85 (0.01) | 0.99 (0) | 2 (0) | 0.82 (0.01) | 93.94 (2.29) |
| Segment | |||||||||||
| Apriori | 3253152 | 0.14 (0) | 0.98 (0) | 2.99 (0) | ∞ | 0.85 (0) | 0.48 (0) | 0.84 (0) | 8.57 (0) | 0.33 (0) | 100 (0) |
| Eclat | 3253152 | 0.14 (0) | 0.98 (0) | 2.99 (0) | ∞ | 0.85 (0) | 0.48 (0) | 0.84 (0) | 8.57 (0) | 0.41 (0) | 100 (0) |
| NICGAR | 12.6 | 0.22 (0.01) | 0.99 (0.01) | 4.91 (0.43) | ∞ | 0.98 (0.01) | 0.97 (0.01) | 1 (0) | 2 (0) | 0.88 (0.01) | 99.76 (0.26) |
| Sonar | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 22.8 | 0.26 (0.03) | 0.89 (0.02) | 7.56 (5.59) | ∞ | 0.81 (0.02) | 0.77 (0.02) | 0.97 (0.01) | 2.01 (0.02) | 0.84 (0.02) | 100 (0) |
| Spambase | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 11.2 | 0.12 (0.02) | 0.95 (0.02) | 73.02 (28.16) | ∞ | 0.9 (0.03) | 0.87 (0.04) | 0.98 (0.01) | 2 (0) | 0.84 (0.05) | 76.75 (31.96) |
| Spectfheart | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 72.2 | 0.13 (0.01) | 0.98 (0) | 29.01 (4.73) | ∞ | 0.92 (0) | 0.92 (0.02) | 1 (0) | 2.03 (0.05) | 0.89 (0.02) | 100 (0) |
| Stock | |||||||||||
| Apriori | 855 | 0.13 (0) | 0.91 (0) | 4.77 (0) | ∞ | 0.88 (0) | 0.76 (0) | 0.96 (0) | 4.16 (0) | 0.44 (0) | 99.48 (0) |
| Eclat | 855 | 0.13 (0) | 0.91 (0) | 4.77 (0) | ∞ | 0.88 (0) | 0.76 (0) | 0.96 (0) | 4.16 (0) | 0.44 (0) | 99.48 (0) |
| NICGAR | 9.2 | 0.28 (0.02) | 0.95 (0.01) | 3.01 (0.18) | ∞ | 0.93 (0.02) | 0.87 (0.01) | 0.99 (0) | 2.07 (0.07) | 0.86 (0.02) | 99.41 (0.23) |
| Stulong | |||||||||||
| Apriori | 89 | 0.31 (0) | 0.93 (0) | 1.22 (0) | ∞ | 0.43 (0) | 0.14 (0) | 0.29 (0) | 3.26 (0) | 0.43 (0) | 99.86 (0) |
| Eclat | 89 | 0.31 (0) | 0.93 (0) | 1.22 (0) | ∞ | 0.43 (0) | 0.14 (0) | 0.29 (0) | 3.26 (0) | 0.43 (0) | 99.86 (0) |
| NICGAR | 4.2 | 0.26 (0.07) | 0.79 (0.03) | 16.82 (19.9) | ∞ | 0.62 (0.08) | 0.64 (0.06) | 0.9 (0.03) | 2.04 (0.09) | 0.78 (0.06) | 84.99 (7.01) |
| Texture | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 17.4 | 0.27 (0.02) | 0.97 (0) | 3.72 (0.39) | ∞ | 0.95 (0) | 0.92 (0.01) | 1 (0) | 2 (0) | 0.82 (0.02) | 95.85 (4.66) |
| Thyroid | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 6.2 | 0.23 (0.07) | 0.93 (0.02) | 14.29 (9.46) | ∞ | 0.8 (0.07) | 0.85 (0.02) | 0.95 (0.05) | 2.02 (0.05) | 0.79 (0.05) | 98.16 (2.3) |
| Wdbc | |||||||||||
| Apriori | 0 | - | - | - | - | - | - | - | - | - | - |
| Eclat | 0 | - | - | - | - | - | - | - | - | - | - |
| NICGAR | 13.6 | 0.27 (0.03) | 0.96 (0.02) | 3.49 (0.39) | ∞ | 0.93 (0.04) | 0.9 (0.03) | 0.99 (0.01) | 2 (0) | 0.84 (0.01) | 98.53 (2.1) |
| Vehicle | |||||||||||
| Apriori | 141107 | 0.14 (0) | 0.95 (0) | 4.18 (0) | ∞ | 0.91 (0) | 0.76 (0) | 0.95 (0) | 6.4 (0) | 0.47 (0) | 100 (0) |
| Eclat | 141107 | 0.14 (0) | 0.95 (0) | 4.18 (0) | ∞ | 0.91 (0) | 0.76 (0) | 0.95 (0) | 6.4 (0) | 0.47 (0) | 100 (0) |
| NICGAR | 12.8 | 0.27 (0.03) | 0.97 (0.01) | 3.46 (0.26) | ∞ | 0.95 (0.02) | 0.92 (0.01) | 1 (0) | 2 (0) | 0.86 (0.02) | 99.98 (0.05) |
| Wine | |||||||||||
| Apriori | 1348 | 0.13 (0) | 0.91 (0) | 5.97 (0) | ∞ | 0.87 (0) | 0.82 (0) | 0.97 (0) | 4.07 (0) | 0.68 (0) | 100 (0) |
| Eclat | 1348 | 0.13 (0) | 0.91 (0) | 5.97 (0) | ∞ | 0.87 (0) | 0.82 (0) | 0.97 (0) | 4.07 (0) | 0.68 (0) | 100 (0) |
| NICGAR | 6.6 | 0.28 (0.01) | 0.94 (0.01) | 2.79 (0.18) | ∞ | 0.9 (0.02) | 0.84 (0.02) | 0.99 (0.01) | 2 (0) | 0.86 (0) | 94.61 (0.85) |
| Vowel | |||||||||||
| Apriori | 235 | 0.14 (0) | 0.98 (0) | 2.97 (0) | ∞ | 0.96 (0) | 0.69 (0) | 0.97 (0) | 3.48 (0) | 0.7 (0) | 100 (0) |
| Eclat | 235 | 0.14 (0) | 0.98 (0) | 2.97 (0) | ∞ | 0.96 (0) | 0.69 (0) | 0.97 (0) | 3.48 (0) | 0.7 (0) | 100 (0) |
| NICGAR | 8.4 | 0.27 (0.11) | 0.95 (0.05) | 5.81 (2.57) | ∞ | 0.92 (0.07) | 0.89 (0.08) | 0.95 (0.09) | 2.02 (0.05) | 0.85 (0.01) | 100 (0) |
